Lagging DNA Strand – Everything You Need To Know
The two strands of the double helix are separated during DNA replication, and each serves as a template for the synthesis of a new complementary strand. However, because the double helix is antiparallel, the two strands are replicated in opposite directions.
The leading strand is the strand that is continuously synthesised in the direction of the advancing replication fork. The lagging strand is the strand that is synthesised in the opposite direction and in short, discontinuous segments.
The lagging strand is a single DNA strand that is replicated in the 5′ – 3′ direction during DNA replication (opposite direction to the replication fork). Okazaki fragments are discontinuous chunks of DNA that are added to the lagging strand.
In this introduction, we will journey into the enthralling world of the lagging DNA strand, discovering its mechanisms, significance, and critical role in the perpetuation of life’s diversity.
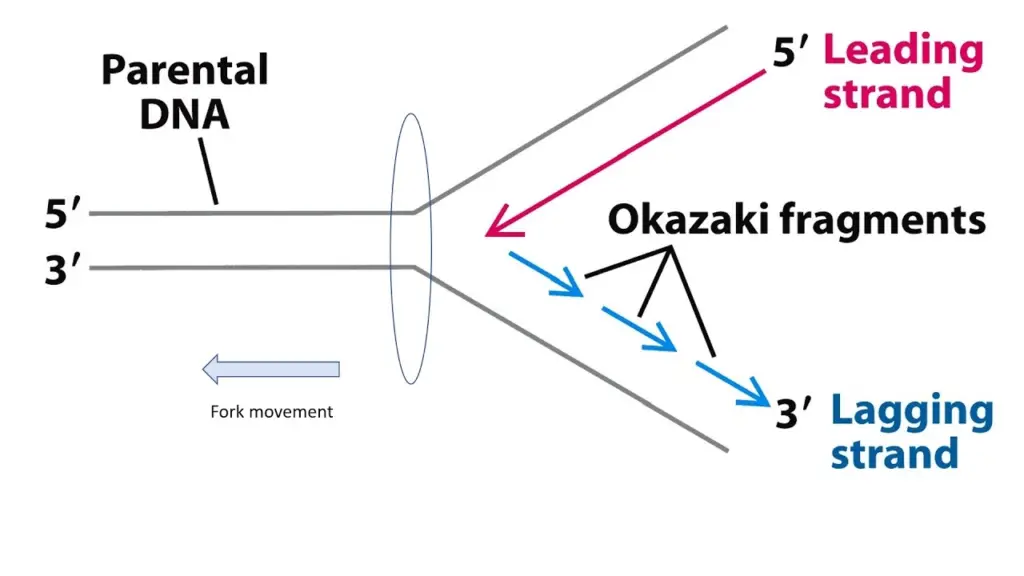
What is the function of the lagging strand in DNA replication?
The function of the lagging strand in DNA replication is mentioned below:
- Duplicating the Code: In DNA replication, the lagging strand is critical in copying the genetic information encoded within the DNA molecule.
- Complexity in Replication: DNA replication is a complex process that involves unwinding the double helix structure and the synthesis of two new strands. While the leading strand is constantly synthesised, the lagging strand faces a special challenge due to its antiparallel orientation.
- Fragmented Replication: The lagging strand is synthesised in small fragments known as Okazaki fragments, which are typically 100 to 200 nucleotides long.
- Primase’s Initiating Role: To begin synthesis on the lagging strand, an enzyme called primase first synthesises an RNA primer. This primer serves as a jumping off point for DNA synthesis.
- Polymerase on the Go: Polymerase III then attaches to the RNA primer and adds complementary nucleotides to form a complementary DNA strand. However, because the lagging strand runs in the opposite direction, the polymerase must “backstitch” away from the replication fork.
- Okazaki Fragment Formation: Okazaki The lagging strand grows in a series of short segments as the replication fork continues to unwind. Each time the helix unwinds, primase adds a new RNA primer, followed by DNA synthesis by polymerase III.
- RNase H and Primer Removal: After a short segment is synthesised, an enzyme called RNase H removes the RNA primer, leaving a gap in the DNA sequence.
- Ligase Seals the Deal: After the RNA primer is removed, a different DNA polymerase, known as DNA polymerase I fills in the gap with complementary nucleotides. Finally, the nicks between adjacent Okazaki fragments are sealed by DNA ligase, resulting in a continuous strand.
- Ensuring Accuracy: Despite the fragmented synthesis, the lagging strand maintains a high level of accuracy during DNA replication. To correct any errors that may occur during polymerization, proofreading mechanisms are in place.
- Collaboration: The lagging and leading strands work in tandem to ensure that the entire DNA molecule is accurately duplicated during cell division, allowing genetic information to be passed down to daughter cells.
How is the synthesis of the lagging strand different from the leading strand during DNA replication?
5 Key Differences in Synthesising the Leading and Lagging Strands During DNA Replication
- Direction of Synthesis:
- Leading Strand: The leading strand is synthesised continuously in the 5′ to 3′ direction, which is the same as the movement of the replication fork.
- Lagging Strand: The lagging strand is synthesised discontinuously in short fragments called Okazaki fragments. Each Okazaki fragment is synthesised in a 5′ to 3′ direction away from the replication fork.
- Primase Activity:
- Leading Strand: The leading strand requires only one RNA primer at the origin of replication to initiate synthesis.
- Lagging Strand: The lagging strand requires multiple RNA primers, one for each Okazaki fragment, to initiate each round of synthesis.
- Polymerase Movement:
- Leading Strand: DNA polymerase continuously moves along the template strand in the 3′ to 5′ direction, allowing for smooth and continuous synthesis.
- Lagging Strand: DNA polymerase moves in a “stop-and-go” manner, creating short stretches of DNA in each Okazaki fragment.
- Synthesis Rate:
- Leading Strand: The leading strand is synthesised at a faster rate due to its continuous process and minimal interruptions.
- Lagging Strand: The lagging strand is synthesised at a slower rate due to the repeated start-and-stop nature of Okazaki fragment synthesis.
- Ligase Involvement:
- Leading Strand: Ligase activity is minimal on the leading strand, as there are no discontinuities to be sealed.
- Lagging Strand: DNA ligase plays a crucial role in sealing the nicks between Okazaki fragments, creating a continuous sugar-phosphate backbone.
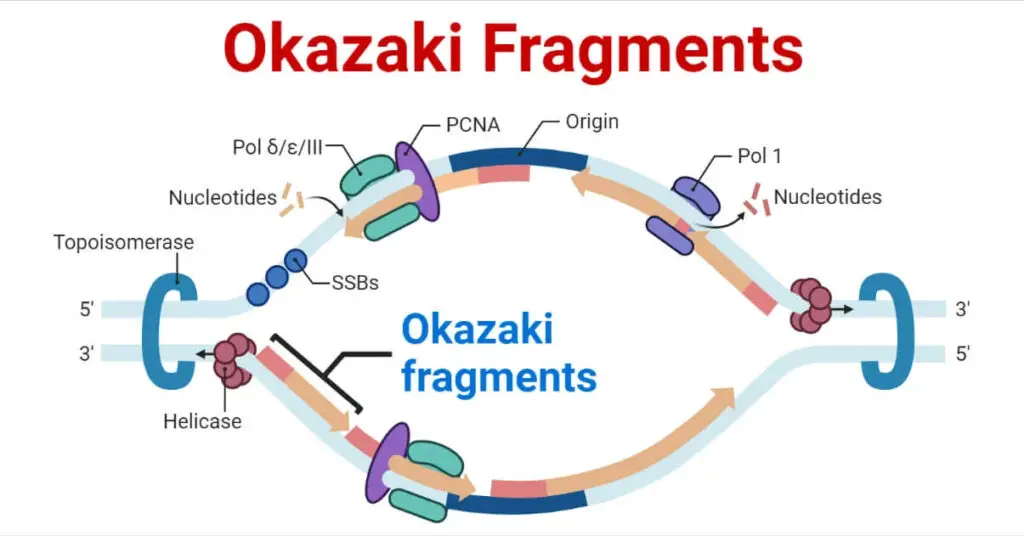
What are Okazaki fragments, and how are they involved in the replication of the lagging strand?
Okazaki fragments are essential for lagging strand replication during DNA replication. This intricate process ensures that genetic information is faithfully duplicated, and these fragments are essential players in this complex molecular dance.
Let’s look at what Okazaki fragments are and how they help with replication:
- Defining Okazaki Fragments: Okazaki fragments are short, discontinuous segments of newly synthesised DNA that are generated during DNA replication on the lagging strand. They were named after the Japanese scientist Reiji Okazaki, who made significant contributions to the understanding of DNA replication with his wife Tsuneko.
- The Replication Process: The process of duplicating a parent DNA molecule to produce two identical daughter DNA molecules is known as DNA replication. It consists of two strands: the leading and lagging strands. The leading strand is continuously synthesized, while the lagging strand is synthesised discontinuously as Okazaki fragments.
- Synthesis of Okazaki Fragments: DNA polymerase synthesises Okazaki fragments on the lagging strand in the opposite direction of the replication fork’s movement. Because DNA polymerase can only add nucleotides in the 5′ to 3′ direction, and the lagging strand runs 3′ to 5′, Okazaki fragment synthesis takes place in short, “backward” stretches.
- RNA Priming: The synthesis of Okazaki fragments begins with the enzyme primase, which produces a short RNA primer. This primer provides the 3′-OH group needed for DNA polymerase to begin DNA synthesis.
- Fragment Elongation: After binding to the RNA primer, DNA polymerase begins adding complementary nucleotides to the growing DNA strand in the 5′ to 3′ direction. As the replication fork progresses, it leaves a trail of short Okazaki fragments behind.
- Removal of RNA Primers: When the DNA synthesis for each Okazaki fragment is completed, a separate enzyme called DNA polymerase I replaces the RNA primers with DNA nucleotides.
- Ligating Fragments: The fragments remain unconnected after the RNA primers are replaced. A DNA ligase enzyme forms phosphodiester bonds to close the gaps between Okazaki fragments, resulting in a continuous lagging strand.
- Completion of Replication: The lagging strand is now a complete copy of the original template DNA after all Okazaki fragments have been synthesised, replaced, and ligated.
Which enzyme is responsible for synthesizing the Okazaki fragments on the lagging strand?
When it comes to the complex dance of DNA replication, several enzymes take centre stage to ensure the process’s accuracy and efficiency.
The DNA polymerase I enzyme is one of these molecular superheroes. This amazing enzyme is in charge of synthesising Okazaki fragments on the lagging strand during DNA replication, which is critical for maintaining the integrity of our genetic code.
Let’s take a look at the fascinating world of DNA replication and see how DNA polymerase I deals with the problem of lagging strand synthesis:
- Setting the Scene: DNA replication is the process by which a cell duplicates its genetic information. The double-stranded DNA molecule unwinds, and each strand serves as a template for the synthesis of a new complementary strand.
- Leading and Lagging Strands: The DNA strands are oriented in opposite directions, with one strand synthesised continuously in the direction of the replication fork (leading strand), and the other strand synthesised in short, discontinuous fragments away from the replication fork (lagging strand) (lagging strand).
- The Okazaki Fragments: These short, newly synthesised DNA fragments on the lagging strand are known as Okazaki fragments, after the Japanese scientist Reiji Okazaki who first described them.
- Rescue by DNA Polymerase I: DNA polymerase I is a versatile enzyme with multiple functions. It plays an important role in lagging strand synthesis by filling gaps between Okazaki fragments.
- Proofreading and Gap Filling: As DNA polymerase I fills the gaps between Okazaki fragments, it also proofreads the newly synthesised DNA. This proofreading function contributes to the accuracy of DNA replication by preventing errors from entering the genetic code.
- RNA Primer Removal: DNA polymerase is used to remove RNA primers. I not only fills in the gaps with nucleotides, but it also removes the RNA primers that were initially laid down to initiate DNA synthesis. This results in a continuous and uniform DNA strand.
- Ligase Lends a Hand: While DNA polymerase I takes care of most of the gaps, DNA ligase steps in to join the Okazaki fragments together, creating a continuous strand on the lagging strand side.
DNA polymerase I plays the role of a meticulous craftsman in the grand performance of DNA replication, meticulously piecing together the Okazaki fragments on the lagging strand. This enzyme contributes to the accurate transmission of genetic information from one generation to the next by filling gaps and proofreading.
How is DNA ligase crucial for completing the synthesis of the lagging strand?
When it comes to DNA replication, the lagging strand plays an important role in ensuring accurate genetic information duplication. However, because the lagging strand is inherently discontinuous, it requires the assistance of several enzymes to complete the synthesis. Among them, DNA ligase stands out for its critical role in completing the replication process.
Here are the five most important roles of DNA ligase in completing the lagging strand synthesis:
- Okazaki Fragments Sealing: The lagging strand is synthesised in short, fragmented segments known as Okazaki fragments. Although DNA polymerase works to lengthen these fragments, gaps remain. As the repair crew, DNA ligase seals the nicks between adjacent Okazaki fragments. DNA ligase ensures a continuous, unbroken strand by catalysing the formation of phosphodiester bonds.
- Repairing the Deoxyribose Phosphate Backbone: The removal of RNA primers during lagging strand synthesis leaves gaps in the DNA backbone. A deoxyribose phosphate moiety is present in these gaps. DNA ligase is critical in repairing this damage. It removes the deoxyribose phosphate and replaces it with a phosphate group, effectively closing the gap and preserving the DNA molecule’s integrity.
- Connecting DNA Fragments: DNA polymerase synthesises DNA in the 5′ to 3′ direction, resulting in the formation of new strands. The lagging strand, on the other hand, is synthesised in a discontinuous manner, resulting in separate DNA fragments. DNA ligase functions as a molecular glue, forming covalent bonds to connect these individual fragments. This ligation process completes the lagging strand synthesis.
- Proofreading and Quality Control: Although DNA replication strives for accuracy, mistakes can occur. Before sealing the nicks, DNA ligase has a built-in proofreading mechanism that ensures the correct bases are paired. This quality control feature aids in the preservation of the genetic code, preventing mutations and potential health problems in the future.
- Ensuring Efficient Replication: DNA ligase enhances the efficiency of DNA replication by swiftly sealing the gaps and nicks in the lagging strand. Without DNA ligase’s action, the gaps could lead to instability in the DNA molecule or even hinder further replication. By rapidly connecting the fragments, DNA ligase ensures a smooth replication process.
What is the significance of the 5′ to 3′ directionality in DNA replication on the lagging strand?
The Significance of 5′ to 3′ Directionality in DNA Replication on the Lagging Strand
- Maintaining Genetic Integrity: DNA replication is a complex process that is required for cell division and the preservation of genetic information. The 5′ to 3′ directionality of DNA replication on the lagging strand is critical in ensuring DNA replication fidelity.
- Leading and Lagging Strands: DNA replication requires two complementary strands: leading and lagging. The leading strand is continuously synthesised in the 3′ to 5′ direction, following the replication fork’s movement. Due to the antiparallel nature of DNA, the lagging strand is synthesised in fragments known as Okazaki fragments.
- Okazaki Fragment Formation: The lagging strand’s 5′ to 3′ directionality results in the discontinuous synthesis of Okazaki fragments. Because DNA polymerase can only add nucleotides to the 3′ end of a growing strand, the lagging strand must be synthesised in the opposite direction as the replication fork.
- Primase and RNA Primers: A short RNA primer is synthesised by an enzyme called primase to start the synthesis of each Okazaki fragment. This primer contains a free 3′ OH group to which DNA polymerase can bind nucleotides. The primer is positioned at the 5′ end of the lagging strand due to the 5′ to 3′ directionality, allowing for efficient DNA synthesis.
- Okazaki Fragment Joining: After each Okazaki fragment is synthesised, the RNA primer is removed by a specialised enzyme, leaving a gap. The gap is then filled with DNA nucleotides by DNA polymerase. Another enzyme, DNA ligase, eventually joins the fragments together. Because of the 5′ to 3′ directionality, DNA ligase can effectively seal the fragments, resulting in a continuous lagging strand.
- Ensure Accuracy: The 5′ to 3′ directionality also contributes to DNA replication accuracy. DNA polymerase can only proofread in the 3′ to 5′ direction, which corresponds to the leading strand’s continuous synthesis. This configuration ensures that nucleotide incorporation errors are efficiently corrected on both strands.
- Base Pairing Consistency: DNA replication is based on complementary base pairing between the template strand and the newly synthesised strand. The 5′ to 3′ directionality of DNA replication on both strands ensures that adenine (A) pairs with thymine (T) and cytosine (C) pairs with guanine (G), preserving the fidelity of the genetic code.
How does the replication machinery handle the repetitive priming process on the lagging strand?
When it comes to DNA replication, the intricate dance of enzymes and proteins is nothing short of a marvel. Among the many intricate steps involved, the repetitive priming process on the lagging strand stands out as a crucial mechanism. Let’s dive into how the replication machinery adeptly handles this process.
- Laying the Groundwork: Primer Synthesis
The replication process begins with helicase unwinding the DNA double helix. This reveals the template strands, including the lagging strand, where synthesis occurs in the opposite direction of fork movement. To begin synthesis, DNA polymerase requires a primer, which is a short RNA strand. Primase, an RNA polymerase, enters the picture to produce these primers at regular intervals along the lagging strand. - Dealing with Repetition: Okazaki Fragments
Due to the unidirectional nature of DNA polymerase synthesis, the lagging strand must be synthesised in short fragments known as Okazaki fragments. Primase synthesises primers repeatedly as the replication fork advances, each of which initiates a new Okazaki fragment. Despite its fragmented nature, this repetitive priming process ensures continuous synthesis of the lagging strand. - Handing over the Baton: Coordinated Enzymatic Efforts
After primase has produced a primer, DNA polymerase takes over. Attaching to the primer-template junction, DNA polymerase begins adding complementary nucleotides in a 5′ to 3′ direction. It eventually reaches the previously synthesised Okazaki fragment as it progresses. This is where the repeated priming process comes into play once more. - RNase H and Primer Removal
To allow for continuous synthesis, the RNA primers in the Okazaki fragments must be removed. This is where RNase H enters the picture. RNase H recognises the primer’s RNA-DNA hybrid structure and degrades the RNA component. This creates a “nick” in the DNA by leaving a gap where the primer used to be. - Filling the Gaps: DNA Polymerase and Ligase
The gaps created by primer removal are then filled by a different DNA polymerase known as DNA polymerase. This enzyme fills gaps in the DNA strand with the appropriate nucleotides. After filling the gap, DNA ligase seals the nick by catalysing the formation of a phosphodiester bond between adjacent DNA fragments, resulting in a continuous strand.
So, here it’s all of the ability of the replication machinery to handle the repetitive priming process on the lagging strand demonstrates the intricate coordination and teamwork of enzymes and proteins. This process ensures the accurate duplication of DNA, a cornerstone of life itself, from the synthesis of primers by primase to the gap-filling abilities of DNA polymerases.
Why is the lagging strand synthesized discontinuously in small fragments, as opposed to the continuous synthesis on the leading strand?
The lagging strand synthesized discontinuously in small fragments, as opposed to the continuous synthesis on the leading strand because:
- Directionality Dilemma: DNA polymerases, the enzymes responsible for DNA replication, can only add new nucleotides to the 3′ end of an existing DNA strand. Because the lagging strand is oriented in the opposite direction, this presents a challenge. To overcome this, the lagging strand must be synthesised in Okazaki fragments, which allow DNA polymerase to work on its 3′ end.
- Leading the Way with the Leading Strand: The leading strand is replicated in the same direction as the replication fork moves. This allows it to be continuously synthesised in a smooth and efficient manner. As the replication fork opens, the DNA polymerase can simply attach to it and replicate the leading strand.
- Forking Complications: The lagging strand, on the other hand, presents a more difficult scenario. As the replication fork advances, the lagging strand is exposed in the opposite direction of the fork movement. This means that, unlike the leading strand, DNA polymerase cannot simply attach and replicate the entire strand in one go.
- Okazaki Fragments to the Rescue: DNA replication employs a clever solution to address this issue. Okazaki fragments are short segments that replicate the lagging strand. At regular intervals on the lagging strand, DNA primase synthesises a short RNA primer. The primers are then extended by DNA polymerase, resulting in Okazaki fragments. Another enzyme, DNA ligase, then stitches these fragments together.
- Ensuring Accuracy: DNA replication is a high-fidelity process for both the leading and lagging strands. The cell can reduce the risk of errors by replicating the lagging strand in fragments. If an error occurs in a single fragment, it is possible to repair it without affecting the entire strand.
- Energetic Efficiency: DNA replication necessitates the use of energy in the form of ATP. Synthesizing the lagging strand in fragments is more energy efficient because it allows the cell to conserve resources by only using energy when necessary.
What are the challenges or issues that might arise during the replication of the lagging strand?
5 Challenges in Replicating the Lagging Strand During DNA Replication
- Okazaki Fragment Formation: The lagging strand replication involves the formation of short DNA fragments known as Okazaki fragments. These fragments must be synthesised correctly in the opposite direction of the replication fork. Coordination of the timely synthesis and joining of these fragments may pose challenges in terms of fidelity and efficiency.
- RNA Priming: The primase enzyme must first synthesise a short RNA primer before DNA polymerase can begin synthesis of the lagging strand. Coordination of primer synthesis and removal after DNA synthesis is difficult because errors can result in gaps or mismatches in the replicated strand.
- Slippage and Replication Errors: Certain DNA sequences’ repetitive nature can cause slippage during replication. This phenomenon can cause nucleotide insertions or deletions, resulting in frameshift mutations. Mismatched base pairing, for example, can occur during replication, necessitating the use of proofreading mechanisms to maintain genomic integrity.
- Replication Fork Stalling: Because of its discontinuous nature, the lagging strand is more prone to replication fork stalling. DNA polymerase can be slowed by obstacles such as DNA damage, secondary structures, or tightly bound proteins. The challenges of restarting stalled replication forks and preventing DNA damage-induced mutations are ongoing.
- Coordination of Enzymes and Factors: DNA polymerase, ligase, helicase, and sliding clamps are among the enzymes and accessory proteins involved in lagging strand synthesis. It is critical that they coordinate their activities and work in the correct order. Incomplete replication, DNA breaks, and inefficient synthesis can all result from disruptions in this coordination.
How does the proofreading mechanism ensure the fidelity of DNA replication on both the leading and lagging strands?
These are how the proofreading mechanism ensure the fidelity of DNA replication on both the leading and lagging strands:
- Leading and Lagging Strands: A Brief Overview
DNA replication consists of two strands: the leading strand, which is continuously synthesised, and the lagging strand, which is synthesised in small fragments known as Okazaki fragments. - DNA Polymerase: The Replication Workhorse
DNA polymerases are enzymes responsible for adding nucleotides to the growing DNA strand during replication. They work hand-in-hand with the proofreading mechanism to minimize errors. - Proofreading Exonuclease Activity
The exonuclease domain is a specialised domain found in DNA polymerases. This domain functions as a proofreader, detecting and correcting errors in the newly synthesised strand. - Detecting Mismatches
As DNA polymerase adds nucleotides to the growing strand, it checks for complementary base pairing. If a mismatch is detected (e.g., A paired with G instead of T), the proofreading exonuclease activity is triggered. - Exonucleolytic Cleavage
When a mismatch is detected, the DNA polymerase changes conformation. This alteration activates the exonuclease domain, which then cleaves the incorrect nucleotide from the growing strand’s 3′ end. - Correcting the Error
After cleaving the incorrect nucleotide, the polymerase returns to its polymerization mode. It then continues to add the correct nucleotide, ensuring proper base pairing. - Continuous Surveillance
The replication process is constantly monitored by the proofreading mechanism. This dynamic interaction between polymerization and proofreading ensures that errors are detected and corrected in real time. - Ensuring Fidelity
The combined action of DNA polymerases and the proofreading mechanism reduces the error rate dramatically during DNA replication. This fidelity is essential for an organism’s genetic integrity. - Role in Disease Prevention
Errors in DNA replication can lead to mutations, which can subsequently contribute to diseases like cancer. The proofreading mechanism acts as a defence mechanism against such detrimental mutations. - Complexity and Evolution
The presence of a proofreading mechanism emphasises the complex nature of DNA replication and the importance of accurately preserving genetic information over evolutionary time.
