What Are Viruses? Structure, Replication, and Host Interactions
Table of Contents
I. Introduction to Viruses
Viruses represent a unique paradigm in the study of microorganisms, characterized by their dependence on host cellular machinery for replication and their remarkable diversity in structure and function. Unlike cellular organisms, viruses lack independent metabolic processes and can only propagate within a suitable host. Their basic structure typically consists of a nucleic acid core—either DNA or RNA—encased in a protective protein shell known as a capsid, often further enveloped by a lipid membrane derived from host cells. The intricacies of viral life cycles, such as entry mechanisms and the molecular interactions with host cell components, reveal how viruses circumvent host defenses to facilitate their replication. For instance, understanding the replication cycle of SARS-CoV-2 elucidates critical processes that can be targeted by antiviral therapies, as illustrated in . This foundational knowledge is crucial for delving into the subsequent themes of virus-host interactions and the implications for public health and disease management.
| Virus Type | Examples | Genome Size (kb) | Host Range | Replication Method |
| DNA Viruses | Herpesviruses, Adenoviruses | 125-230 | Animals and Humans | Nuclear |
| RNA Viruses | Influenza, HIV | 8-30 | Animals, Humans, Plants | Cytoplasmic |
| Retroviruses | HIV, HTLV | 7-10 | Humans | Reverse Transcription |
| Bacteriophages | T4, Lambda | 10-200 | Bacteria | Lytic or Lysogenic |
Viruses Overview Statistics
A. Definition of Viruses
Viruses can be defined as acellular entities composed of nucleic acids—either DNA or RNA—encased in a protein coat, known as a capsid, and in some cases, a lipid envelope. These microscopic pathogens lack the cellular machinery necessary for replication and metabolism, making them obligate parasites that depend entirely on host cells for propagation. Their structural simplicity belies their complexity in behavior and interaction with host organisms. For instance, the viral life cycle encompasses several critical phases, including attachment, entry, replication, and release, each requiring specific interactions with host cell machinery. Understanding these intricate processes is crucial for developing effective antiviral strategies. The representation of the viral life cycle, as seen in , highlights the various stages of viral replication and the interplay between viral proteins and host cellular processes, providing valuable insights into how viruses exploit host systems for their own replication and how interventions can disrupt these interactions.
| Characteristic | Description |
| Type | Viruses can be classified by their type, such as DNA or RNA viruses. |
| Size | Viruses generally range from 20 to 300 nanometers in diameter. |
| Replication | Viruses replicate only inside a host cell by hijacking the host’s cellular machinery. |
| Structure | Viruses consist of genetic material surrounded by a protein coat and sometimes an envelope. |
| Host Interaction | Viruses require a living host to reproduce and can infect a wide variety of organisms. |
Virus Characteristics
B. Are Viruses Living Organisms?
The classification of viruses as living organisms remains a contentious topic in biological sciences due to their unique characteristics and dependency on host cells for replication. Unlike cellular organisms, viruses lack independent metabolic processes, cellular structures, and the ability to reproduce outside a host organism. This dependence raises significant questions about their status as living entities. They exist in a state of biological limbo, displaying characteristics typical of living organisms, such as the ability to evolve and adapt, while simultaneously exhibiting non-living traits by requiring a host cells machinery for reproduction. The intricate interactions during the viral replication cycle, as depicted in the life cycle image , emphasize the complex relationship between viruses and host cells. This relationship highlights the critical nature of understanding viruses not merely as pathogens but as entities that embody the threshold between living and non-living systems, prompting ongoing debate regarding their classification within biological taxonomy.
| Characteristic | Living Organisms | Viruses | Non-Living Entities |
| Cell Structure | Composed of one or more cells | Consist of a protein coat and genetic material, no cellular structure | No cellular structure |
| Metabolism | Capable of metabolic processes | No metabolic processes; rely on host cells | No metabolic processes |
| Growth and Reproduction | Grow and reproduce independently | Cannot grow or reproduce independently; require host cells | Do not grow or reproduce |
| Response to Stimuli | Respond to environmental stimuli | Do not respond to stimuli; can only affect host cells | Do not respond to stimuli |
Comparison of Characteristics of Living and Non-Living Entities
II. Structure of Viruses
Understanding the structure of viruses is crucial for deciphering their mechanisms of infection and replication. Viruses exhibit a variety of structural forms, generally classified into two categories: enveloped and non-enveloped. The viral envelope, composed of a lipid bilayer derived from the host cell membrane, encases vital components such as the viral nucleic acid and proteins. In contrast, non-enveloped viruses possess a protective protein shell, or capsid, that safeguards their genetic material. These structural elements play pivotal roles in the viral life cycle, particularly in the stages of attachment and entry into host cells. For instance, the image illustrating the life cycle of a viral particle highlights critical components like the spike protein and ssRNA, demonstrating their functions in initiating infection and facilitating replication (). Therefore, a comprehensive understanding of viral structures enhances our insight into their interactions with host organisms and informs the development of targeted antiviral strategies.
| Virus Name | Type | Structure | Size (nm) |
| Influenza Virus | RNA Virus | Segmented RNA genome surrounded by a protein coat and lipid envelope | 80-120 |
| HIV (Human Immunodeficiency Virus) | Retrovirus | RNA genome encapsulated in a protein core, surrounded by a lipid envelope | 100-120 |
| COVID-19 (SARS-CoV-2) | RNA Virus | Single-stranded RNA with a helical nucleocapsid surrounded by a lipid bilayer | 60-140 |
| Tobacco Mosaic Virus | RNA Virus | Rigid rod structure made of protein subunits surrounding RNA | 15-20 |
| Bacteriophage T4 | DNA Virus | Icosahedral head and a tail that injects DNA into host bacteria | 225 |
Virus Structure Comparison
A. Viral Genome (DNA vs. RNA)
The genetic material of viruses can be categorized into two main types: DNA and RNA, each influencing the viruss replication strategy and interaction with host cells. DNA viruses, characterized by their double-stranded structure, tend to exhibit more stability and pose a lower mutation rate during replication compared to their RNA counterparts. Conversely, RNA viruses, which may be single-stranded or double-stranded, replicate with higher fidelity but are prone to rapid mutations, enabling swift evolutionary adaptation. This distinction significantly impacts the viral life cycle and the development of antiviral strategies, as highlighted in the schematic representation of the viral life cycle that depicts the stages of RNA virus replication and interaction with host machinery . Understanding the differences in viral genome types is crucial for developing targeted therapies and vaccines, as each type elicits distinct immune responses and presents unique challenges for eradication, making the study of viral genomics essential in virology and public health.
| Virus Type | Examples | Genome Structure | Replication Location | Mutation Rate |
| DNA Virus | Herpes Simplex Virus, Varicella-Zoster Virus | Double-stranded or single-stranded DNA | Nucleus | Lower |
| RNA Virus | Influenza Virus, SARS-CoV-2, HIV | Single-stranded or double-stranded RNA | Cytoplasm | Higher |
| Reverse Transcribing Virus | HIV, Hepatitis B Virus | Single-stranded RNA or Double-stranded DNA (via reverse transcription) | Nucleus and Cytoplasm | Moderate to High |
Viral Genome Comparison: DNA vs. RNA Viruses
B. Capsid and Envelope
The capsid and envelope are critical structural components that significantly influence viral functionality and interactions with host cells. The capsid, composed of protein subunits called capsomers, encases the viral genetic material, providing protection and facilitating the delivery of its contents into host cells during infection. Meanwhile, the envelope, derived from the host cell membrane, contains viral glycoproteins essential for receptor binding and entry. This dual-structure allows viruses to evade the immune system while enhancing their infectivity. Understanding these components is crucial for developing antiviral strategies, as demonstrated in the detailed lifecycle of viral particles, which illustrates how the assembling capsid interacts with the envelope during virion release from host cells . The interplay between the capsid and envelope not only determines the viruss stability and mode of transmission but also influences its susceptibility to antiviral treatments, underscoring the importance of these structures in viral pathogenesis and therapeutic interventions.
| Virus Name | Capsid Type | Envelope | Genome Type |
| HIV | Icosahedral | Yes | RNA |
| Influenza | Helical | Yes | RNA |
| Tobacco Mosaic Virus | Helical | No | RNA |
| Ebola | Filamentous | Yes | RNA |
| Bacteriophage T4 | Complex | No | DNA |
| Herpes Simplex Virus | Icosahedral | Yes | DNA |
| Poliovirus | Icosahedral | No | RNA |
| SARS-CoV-2 | Helical | Yes | RNA |
Viruses Structure: Capsid and Envelope Types
III. Replication of Viruses
The replication of viruses is a remarkably intricate process that occurs within host cells, significantly shaping viral pathogenesis and infection dynamics. Upon entering a susceptible host cell, viruses hijack the cellular machinery to replicate their genetic material and synthesize viral proteins. The viral life cycle begins with attachment and entry, followed by the uncoating of the viral genome, which can be DNA or RNA. Once released, the viral genome is replicated and translated into proteins needed for assembly. This process is illustrated in detail in , which outlines the sequential steps of the replication cycle for SARS-CoV-2, demonstrating how viral components are synthesized and assembled within specialized cellular structures. The hosts cellular environment plays a critical role in this replication, as various host proteins can either assist or impede the viral lifecycle. Understanding these mechanisms is crucial in the ongoing effort to develop effective antiviral therapies that can target specific stages of replication.
| Virus | Replication Time (hours) | Method of Replication |
| Influenza Virus | 6-8 | Negative-sense single-stranded RNA |
| HIV (Human Immunodeficiency Virus) | 24-36 | Positive-sense single-stranded RNA |
| Bacteriophage T4 | 30 | Double-stranded DNA |
| Ebola Virus | 6-10 | Negative-sense single-stranded RNA |
| SARS-CoV-2 (COVID-19) | 8-12 | Positive-sense single-stranded RNA |
Virus Replication Times and Methods
A. Lytic vs. Lysogenic Cycle
The mechanisms by which viruses propagate within host cells can be categorized into two primary cycles: the lytic cycle and the lysogenic cycle, each playing a distinct role in viral life histories and interactions with host organisms. In the lytic cycle, viral replication occurs rapidly, culminating in the death of the host cell through lysis, thus releasing a new generation of virions to infect additional cells. Conversely, the lysogenic cycle allows viral DNA to integrate into the host genome, remaining dormant as a provirus until it is triggered to enter the lytic phase, thereby enabling viral propagation without immediate harm to the host. This nuanced understanding of viral strategies is essential for comprehending their evolutionary adaptations and pathogenic potential. The complexity of these cycles is highlighted in , which illustrates the stages of viral RNA synthesis and structure formation, thereby providing insight into how these mechanisms enable viruses like SARS-CoV-2 to efficiently replicate and interact with host cells.
| Cycle Type | Description | Example Virus | Time to Cell Lysis (hours) | Symptoms |
| Lytic | Virus replicates immediately, resulting in the destruction of the host cell. | T4 Bacteriophage | 1-3 | Rapid onset of symptoms; may include inflammation and cell death. |
| Lysogenic | Virus integrates its genetic material into the host’s genome, remaining dormant until triggered. | Lambda Bacteriophage | Varies (can remain dormant for years) | Delayed onset of symptoms; may trigger later due to stress or other factors. |
Lytic vs. Lysogenic Cycle Overview
B. Viral Entry Mechanisms
Understanding viral entry mechanisms is crucial for elucidating how viruses successfully penetrate host cells to initiate infection. Viruses employ a variety of strategies to attach and enter cells, often utilizing specific receptors on the hosts surface. For instance, enveloped viruses such as SARS-CoV-2 utilize the spike protein to bind to the ACE2 receptor, a key initial step that facilitates fusion of the viral envelope with the host cell membrane. This complex interaction underscores the specificity and adaptability of viral entry processes. Additionally, the role of host cell machinery is pivotal, as it aids in uncoating and releasing viral genetic material into the cytoplasm. The intricacies of these mechanisms, including receptor-mediated endocytosis or membrane fusion, are vital for viral replication and propagation. The processes involved are illustrated in , which highlights the entire replication cycle, further contextualizing the importance of viral entry in the broader scope of viral pathogenesis and therapeutic intervention.
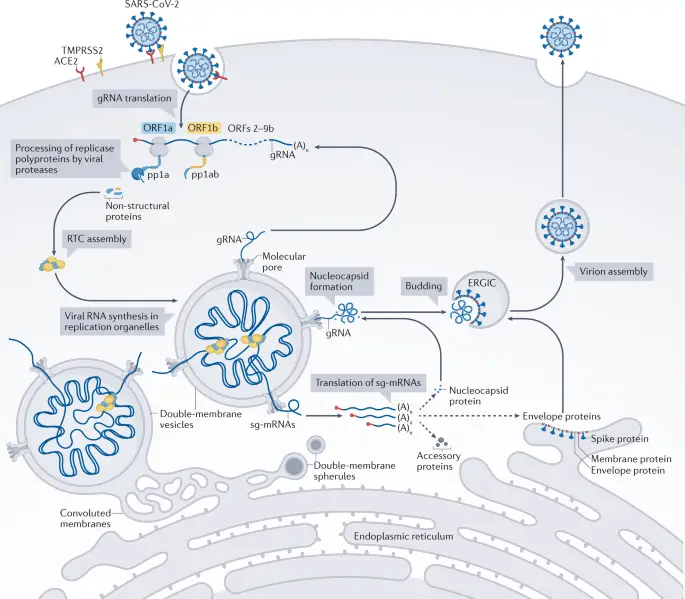
Image : Molecular Replication Cycle of SARS-CoV-2 (The image illustrates the molecular replication cycle of SARS-CoV-2, detailing the various steps involved in viral RNA synthesis and structure formation within host cells. It highlights key components such as the translation of genomic RNA (gRNA), processing of replicase polyproteins by viral proteases, the formation of non-structural proteins, and the assembly of virions in structures like the ER-Golgi intermediate compartment (ERGIC). Additionally, it depicts the role of replication organelles, nucleocapsid formation, and the translation of subgenomic mRNAs (sg-mRNAs), ultimately showcasing the complex interactions at the cellular level that enable the virus to propagate within a host organism.)
IV. Viruses and Their Hosts
The intricate relationship between viruses and their hosts is characterized by a dynamic interplay that impacts not only viral replication but also host cellular functions. Understanding this relationship is crucial, as it reveals the mechanisms by which viruses exploit host machinery to facilitate their life cycles, a process intricately depicted in the viral life cycle diagram. This diagram illustrates essential steps like viral entry, uncoating, and the replication of viral RNA, emphasizing how viruses hijack host cellular processes (see ). Additionally, the interactions between viral proteins and host cell receptors dictate the efficiency of infection and the subsequent immune response, which can vary significantly across different hosts. Such variability underscores the evolutionary arms race between viruses and host defenses, highlighting the need for ongoing research into therapeutic interventions. As viruses adapt, understanding these interactions becomes increasingly vital for developing effective antiviral strategies capable of mitigating viral spread and enhancing public health outcomes.
| Virus | Host | Transmission | Symptoms | Prevention |
| HIV | Humans | Sexual contact, blood transfusion, sharing needles | Immune system deterioration, opportunistic infections | Safe sex practices, antiretroviral therapy |
| Influenza | Humans, Birds, Pigs | Respiratory droplets, surfaces | Fever, cough, body aches | Annual vaccination, hand hygiene |
| COVID-19 (SARS-CoV-2) | Humans | Respiratory droplets, aerosols, contact with contaminated surfaces | Fever, cough, difficulty breathing, loss of taste/smell | Vaccination, wearing masks, physical distancing |
| Zika Virus | Humans, Mosquitoes | Mosquito bites, from mother to fetus | Fever, rash, joint pain, conjunctivitis | Mosquito control, avoiding mosquito bites |
| Ebola Virus | Humans, Fruit Bats | Direct contact with bodily fluids, infected animals | Fever, severe headache, vomiting, bleeding | Avoiding contact with infected individuals, vaccination |
Virus-Host Interactions
A. Host Specificity
Host specificity is a fundamental concept in understanding the interactions between viruses and their potential hosts, impacting both viral evolution and disease transmission. Each virus exhibits a preference for certain host species, largely determined by the molecular compatibility between viral surface proteins and host cell receptors. For instance, the specificity of the HIV virus for human T-cells stems from its envelope proteins binding precisely to the CD4 receptor found on those immune cells. This host-virus interaction not only dictates the range of species a virus can infect but also plays a crucial role in zoonotic transfers, where viruses jump from animals to humans, often leading to outbreaks of novel diseases. The implications of host specificity extend to vaccine development and therapeutic interventions, as understanding these interactions can enhance strategies to mitigate viral infections. As illustrated in [insert image reference], the intricate molecular relationships involved underscore the critical nature of host specificity in the broader context of viral behavior and ecology.
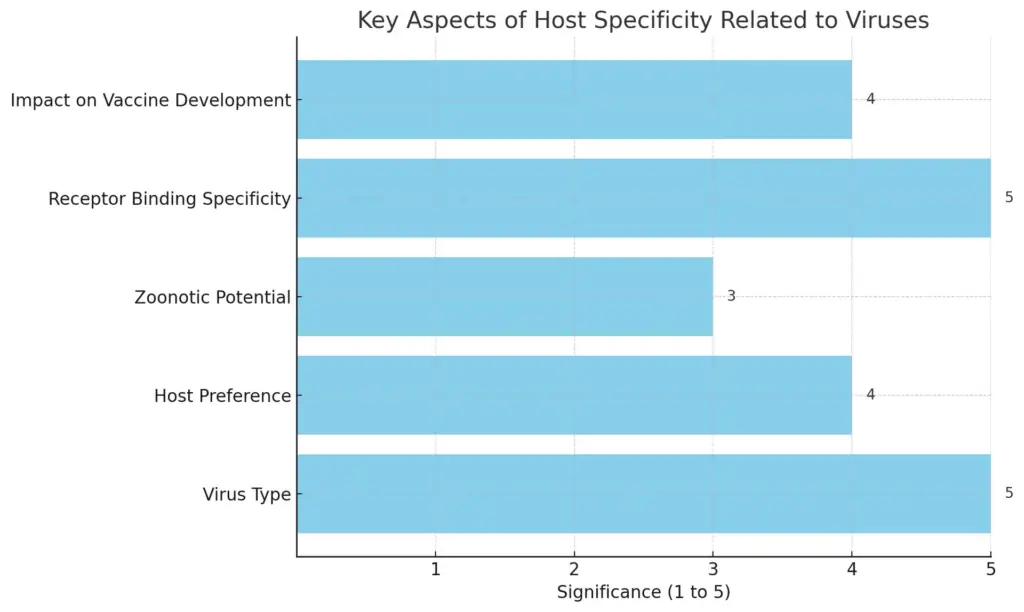
This bar chart illustrates the key aspects of host specificity related to viruses, depicting the significance of various factors on a scale of 1 to 5. The factors include Virus Type, Host Preference, Zoonotic Potential, Receptor Binding Specificity, and Impact on Vaccine Development. Each bar represents a qualitative measure of significance in regards to how these aspects influence virus behavior and vaccine challenges.
B. Zoonotic Viruses
Zoonotic viruses, which are capable of crossing species barriers from animals to humans, represent a significant threat to public health and are a prime example of the intricate relationships between viruses, their hosts, and the environment. The structure of zoonotic viruses often influences their transmissibility and ability to adapt to new hosts, which can lead to outbreaks and pandemics. For instance, viruses such as HIV, influenza, and SARS-CoV-2 have demonstrated the capacity to spill over from wildlife or domestic animals to humans, highlighting the complexity of viral replication mechanisms in diverse biological systems. This adaptability is largely facilitated by mutations in viral genomes that allow for better interaction with host cellular machinery. Furthermore, human behaviors—like encroachment into wildlife habitats and global travel—enhance the likelihood of such interspecies transmission. Understanding the dynamics of zoonotic viruses is crucial for developing effective surveillance and preventive measures against emerging infectious diseases, as illustrated in .
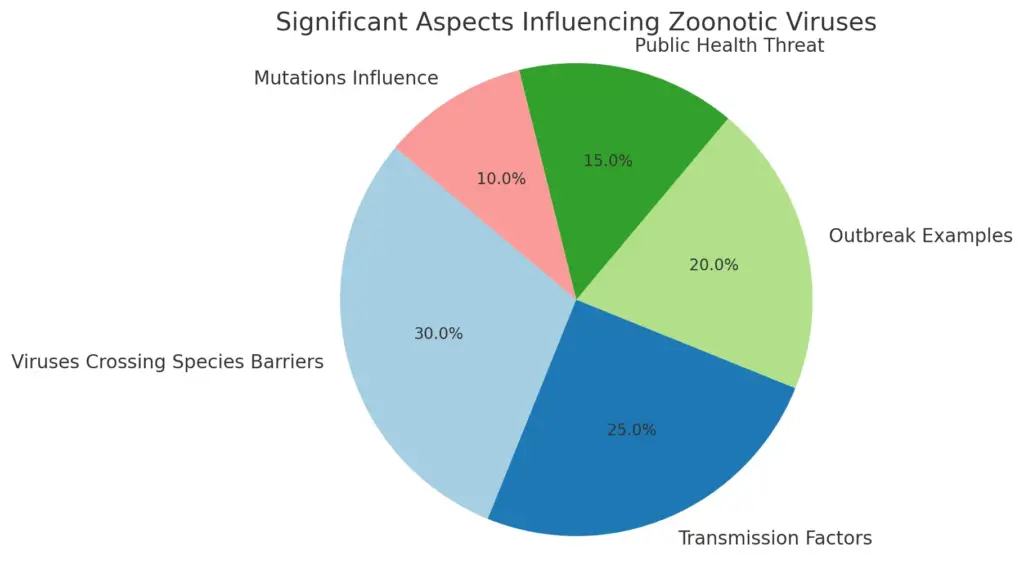
This pie chart represents the significant aspects influencing zoonotic viruses, highlighting the contribution of viruses crossing species barriers, transmission factors, notable outbreak examples, public health threats, and the impact of mutations on viral adaptability. The values indicate the percentage significance of each aspect based on key points extracted from the research.
V. Conclusion
In conclusion, understanding the intricate nature of viruses—including their structure, modes of replication, and interactions with host cells—is crucial for advancing virology and public health strategies. As the ongoing challenges posed by viral infections highlight the importance of research, insights gained from studies of viral life cycles and their molecular dynamics can inform the development of effective antiviral treatments and vaccines. For instance, the detailed schematic representation of the SARS-CoV-2 life cycle presented in effectively underscores the various stages and components critical to viral propagation within host cells, allowing for targeted therapeutic approaches. Additionally, recognizing the complex interplay between viral proteins and host cell mechanisms, as illustrated in , emphasizes the need for comprehensive strategies that address both prevention and treatment. Ultimately, the continuous exploration of viruses, as reflected in their multifaceted interactions, will provide essential knowledge for combating future viral outbreaks and protecting global health.
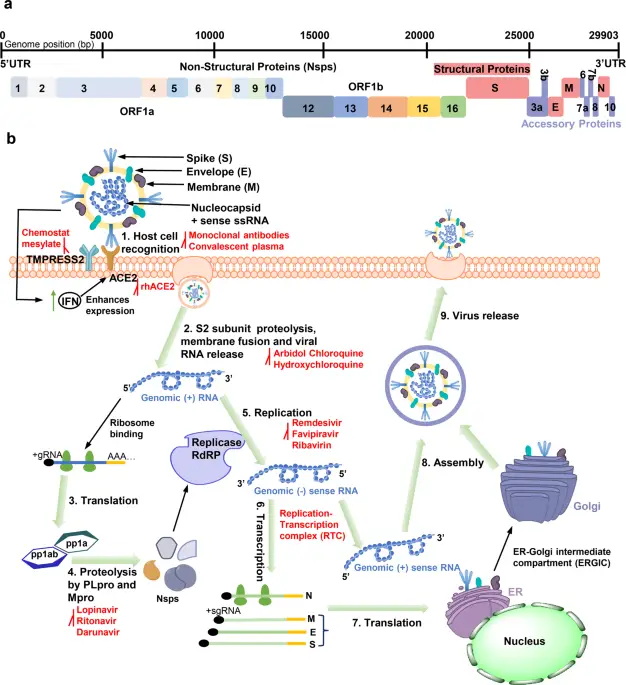
Image : Schematic of SARS-CoV-2 Viral Life Cycle and Antiviral Strategies (The image provides a detailed schematic representation of the SARS-CoV-2 virus life cycle, illustrating critical steps involved in viral infection and replication. It details key processes such as host cell recognition, proteolysis, membrane fusion, and RNA replication. Each step is labeled numerically and includes references to antiviral agents like Remdesivir and chloroquine, emphasizing their roles in the viral life cycle. Additionally, the diagram highlights structural proteins (like Spike and Nucleocapsid) and non-structural proteins, alongside their interactions and processes occurring at the cellular level, contributing to a comprehensive understanding of viral biology and the potential pathways for therapeutic intervention.)
A. Summary of key points
In summarizing the key points of viral structure, replication, and host interactions, it is essential to highlight the multifaceted nature of viral life cycles and their intricate relationships with host cells. Viruses, characterized by their unique structural components, such as the capsid and envelope proteins, utilize host cellular mechanisms for replication, demonstrating a remarkable adaptability that complicates therapeutic interventions. The life cycle of a virus encompasses crucial phases including attachment, entry, uncoating, replication, and assembly, each representing potential targets for antiviral strategies, as depicted in the schematic of the viral life cycle . Moreover, understanding the protein-protein interactions between viral and host proteins is critical for developing effective antiviral therapies and mitigating pathogenic effects, as illustrated in the diagram showcasing these dynamic interactions and therapeutic intervention points . Ultimately, the complexity of virus-host dynamics underscores the necessity for continued research in virology to inform public health strategies and therapeutic advancements.
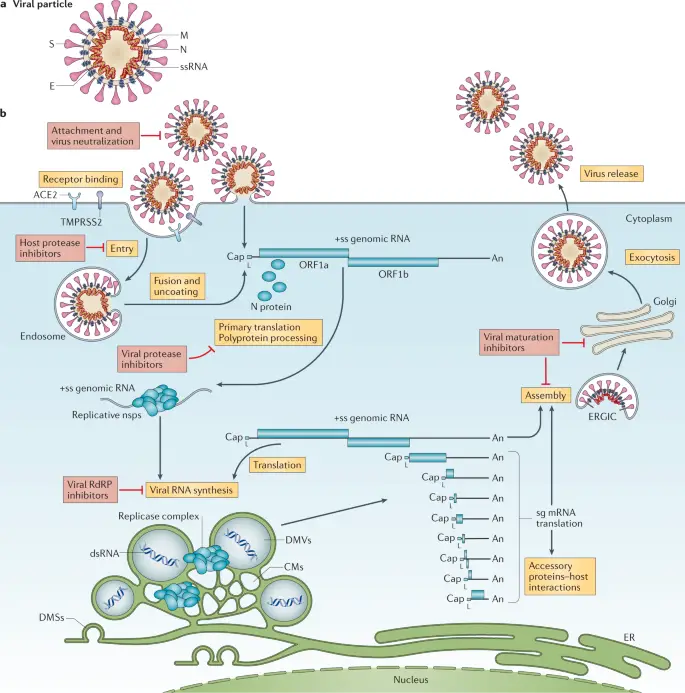
Image : Viral Life Cycle: Overview of Entry, Replication, and Release Mechanisms (The image illustrates the interactions involved in viral-host protein-protein interactions (PPI) and the role of antiviral therapeutics. It features a central node labeled ‘Viral-Host PPI,’ surrounded by various clusters representing protein interactions in different contexts. The left side depicts the virus entering the host cell, while the right side shows the infected cell. Arrows indicate pathways and processes illustrated by scissors symbolizing intervention points for antiviral therapeutics, which are also depicted at the bottom of the image in pill form. This diagram serves to explain the complex dynamics between viruses and host cells and highlights potential targets for antiviral drugs.)
REFERENCES
- Charles Janeway. ‘Janeway’s immunobiology.’ Garland Science, 1/1/2008
- Ying Kong. ‘Tuberculosis Host-Pathogen Interactions.’ Jeffrey D. Cirillo, Springer Nature, 9/10/2019
- Wang-Shick Ryu. ‘Molecular Virology of Human Pathogenic Viruses.’ Academic Press, 3/30/2016
- Jennifer Louten. ‘Essential Human Virology.’ Academic Press, 5/28/2022
- Patrick Materatski. ‘The Application of Viruses to Biotechnology.’ Carla Varanda, MDPI AG, 12/21/2021
- Alistair McCleery. ‘An Introduction to Book History.’ David Finkelstein, Routledge, 3/13/2006
Image References:
- Image: Molecular Replication Cycle of SARS-CoV-2, Accessed: 2025.https://media.springernature.com/m685/springer-static/image/art%3A10.1038%2Fs41580-021-00432-z/MediaObjects/41580_2021_432_Fig1_HTML.png
- Image: Schematic of SARS-CoV-2 Viral Life Cycle and Antiviral Strategies, Accessed: 2025.https://media.springernature.com/m685/springer-static/image/art%3A10.1038%2Fs41392-022-00884-5/MediaObjects/41392_2022_884_Fig1_HTML.png
- Image: Viral Life Cycle: Overview of Entry, Replication, and Release Mechanisms, Accessed: 2025.https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41579-020-00468-6/MediaObjects/41579_2020_468_Fig1_HTML.png
