Methods of Study in Microbiology and Virology
Table of Contents
I. Introduction to Microbial Research
Microbial research is important to understand how microorganisms interact with their surroundings, which is key for progress in health, farming, and biotechnology. This field includes various methods, such as traditional culture techniques, molecular methods, and studying microbial communities in their environments. These approaches are vital for grasping how microbes behave. As researchers create new diagnostic tools, like those mentioned in [citeX], they improve our ability to quickly and accurately detect pathogens, leading to better clinical results and public health measures. Moreover, the use of next-generation sequencing and bioinformatics in microbial studies has changed our understanding of microbial genetics and diversity. This combination of technologies enables scientists to investigate the large genetic variety in microbial communities, revealing connections and functions that were not known before. This fast development in techniques creates new research opportunities, showing the need for constant updates in methods to keep up with scientific progress. It also stresses the increasing need for teamwork across different fields to solve complex microbial problems. Therefore, knowing the background and basic techniques of microbial research—drawing from both classic studies and recent advancements—is crucial for dealing with today’s challenges and innovations in microbiology and virology. By valuing both history and current knowledge, researchers can better steer the future of microbial sciences and use its benefits for society.
| Method | Usage Percentage | Description |
| Culturing | 45 | Traditional method of growing microbes in controlled environments. |
| Molecular Techniques | 30 | Techniques like PCR and gene sequencing for identifying and studying microorganisms. |
| Microscopy | 15 | Visual examination of microbes using optical instruments. |
| Bioinformatics | 10 | Use of software tools to analyze biological data, particularly in genomics. |
Microbial Research Methods and Their Usage Statistics
A. The Importance of Studying Bacteria & Viruses
Studying bacteria and viruses is important for knowing their roles in health, disease, and biotechnology, especially as the microbial world gets more complicated. Researchers look at these microorganisms to better understand the complex interactions that happen in ecosystems and within human hosts. This research helps us improve our basic knowledge and create new treatment methods and public health actions that can greatly enhance health outcomes. For example, the research on ozone as a biocidal agent shows how well it can deactivate germs like E. coli and Salmonella, which points to its important use in food safety and sanitation that helps prevent outbreaks (Anderson et al.). Additionally, learning more about viral structures is essential for improving vaccine development, especially given that the wide variety of viruses creates ongoing challenges for both diagnostics and treatments. Various studies demonstrate that looking at the past of virology reveals important technological progress that has influenced our current methods in microbiology and virology. This highlights how crucial it is to keep researching these areas. Ongoing research is key for finding new ways to deal with future health crises, especially since new pathogens are a big threat to global public health. By staying up to date with the changes in bacteria and viruses, we can better get ready to tackle new issues and utilize the potential advantages these microorganisms might provide.
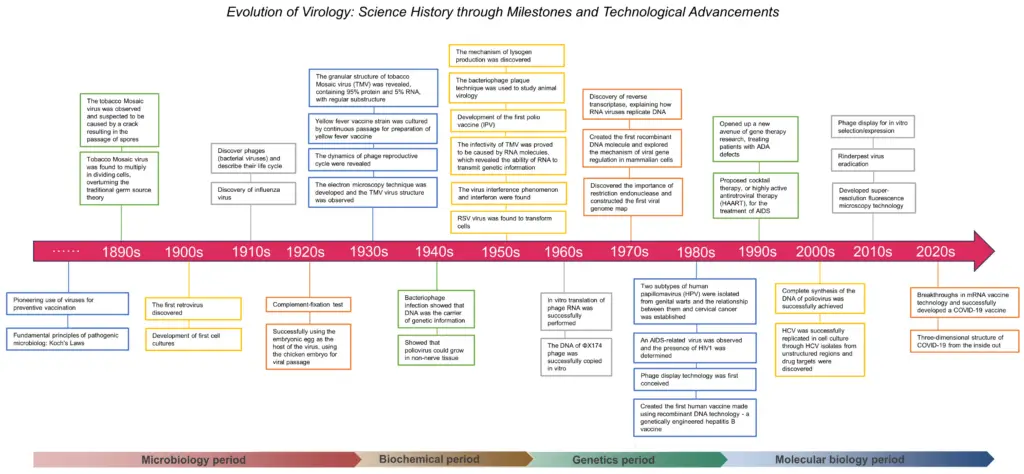
IMAGE – Timeline of the Evolution of Virology: Key Milestones and Technological Advances from the 1890s to the 2020s. (The image is a timeline illustrating the evolution of virology from the 1890s to the 2020s. It highlights significant milestones and technological advancements in the field, categorized into microbiology, biochemistry, genetics, and molecular biology periods. Each decade features key discoveries, such as the first observation of the tobacco mosaic virus, the development of vaccines, and the advent of mRNA vaccine technology. These events showcase the progression of knowledge in virology and its impact on medical science, emphasizing the interactions between viruses and hosts, methodologies like cell cultures, and the molecular foundation of viral genetics. This timeline serves as a valuable resource for understanding the historical context and advancements in virology, relevant for academic reference in fields such as microbiology, molecular biology, and vaccine research.)
B. Key Questions Scientists Seek to Answer
In the complex fields of microbiology and virology, scientists face key questions that shape research and practical use across many areas. A major question is about the link between pathogens and their hosts, which is crucial for creating effective treatments and prevention methods for infectious diseases. For example, the MetaSUB Consortium shows that using interdisciplinary methods like metagenomics is important for finding microbial communities in cities, shedding light on how diseases spread and how antibiotic resistance develops (Consortium MI). This knowledge has practical effects on public health and environmental policies. Moreover, NASA’s environmental health plan raises important questions about how microbes survive in space, broadening the study of virology beyond Earth and prompting scientists to think about life’s potential in other parts of the universe (N/A). Various advanced techniques like imaging and genomic sequencing help researchers gain detailed insights into how microbes act and interact at a molecular level. This advancement in technology keeps the knowledge in the field growing and changing, leading to new findings and a better understanding of microbes. The importance of diagnostic methods highlights the essential role of accurate techniques in addressing these important scientific questions. By concentrating on these questions, scientists not only push the limits of microbiological research but also set the stage for advancements that can greatly enhance health, safety, and quality of life.
| Question | Relevance | Current research focus |
| What are the mechanisms of antibiotic resistance in bacteria? | Understanding resistance can lead to better treatment options and new antibiotics. | Investigating genetic mutations and horizontal gene transfer. |
| How do viruses interact with host cells? | Insights can inform vaccine development and antiviral therapies. | Studying viral entry, replication, and immune evasion strategies. |
| What factors influence the spread of infectious diseases? | Identifying these factors can help control outbreaks and improve public health responses. | Analyzing environmental, social, and biological determinants. |
| How can microbiomes affect human health? | Understanding microbiomes can improve treatment for diseases like obesity and diabetes. | Exploring the links between gut microbiota and metabolic health. |
| What is the role of arthropods in virus transmission? | Targeting vectors may reduce the incidence of viral diseases. | Researching vector biology and control strategies. |
Key Questions in Microbiology and Virology Research
II. Methods for Studying Bacteria
The study of bacteria uses many methods, each designed to reveal different parts of microbial life and behaviors. Techniques like molecular biology have greatly changed our understanding of bacterial genetics, allowing for a more in-depth look at gene functions and regulation that is crucial for microbial survival and adjustment. Furthermore, methods to measure biochemical materials, like proteins and nucleic acids, greatly improve our ability to analyze bacterial behaviors, interactions, and their roles in various ecosystems, helping researchers decode complex microbial groups (Bauer et al.). Recently, new methods to inactivate foodborne germs are being examined, highlighting the real-world applications of these powerful techniques. For example, using ozone to kill bacteria like E. coli and Salmonella enteritidis shows how research can impact public health, stressing the need to understand how pathogens act in different conditions and situations faced in the real world (Anderson et al.). These techniques not only add to basic microbiological knowledge but also tackle urgent global health issues, such as food safety and antibiotic resistance. The continuous improvement of these methods leads to new ways to understand bacterial life, which can help create better treatments and control infections, critical in the global fight against infectious diseases.
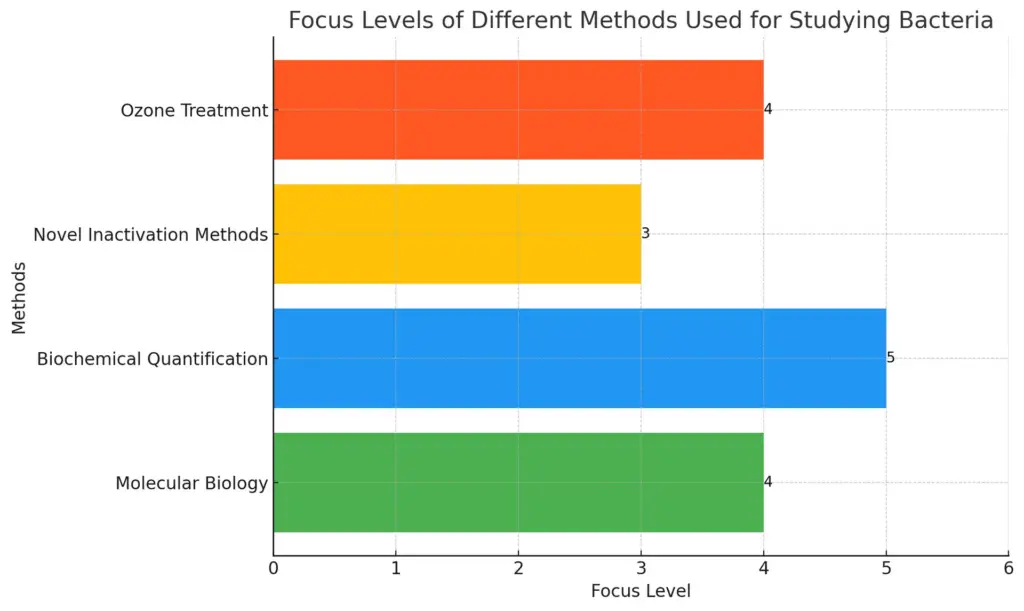
This bar chart illustrates the different methods used for studying bacteria, highlighting the emphasis on molecular biology, biochemical quantification, novel inactivation methods, and the specific case of ozone treatment against pathogens. Each value represents the importance or focus level of each method based on the insights from the paragraph.
A. Microscopy Techniques (Light, Electron)
Microscopy methods, mainly light microscopy and electron microscopy, are key techniques for seeing microorganisms in microbiology and virology. They are vital in many biological research areas. Light microscopy is useful for viewing live samples, as it helps researchers observe biological functions in real-time. This technique gives a broad view of cell structures because it has lower magnification, which is good for spotting larger parts of cells and tissues. Still, it struggles with showing fine details, especially when looking at tiny viral particles that are smaller than what visible light can resolve. This drawback can make it tough to fully understand viral shapes. On the other hand, electron microscopy provides much higher resolutions, allowing for the examination of complex viral structures and their interactions with host cells in more detail. Using these microscopy methods is very important because they help identify pathogens and improve our overall understanding of their biology. This is especially true for viral infections like those from herpes simplex virus (HSV), where imaging works alongside molecular diagnostics like Real-time PCR, creating a more complete way to study viral disease progression. This combination of imaging and molecular techniques is essential for improving diagnosis and treatment methods. Additionally, the importance of these microscopy techniques is illustrated in a diagram showing virus detection through different lab methods, highlighting how imaging and diagnostics relate. This better understanding helps researchers and doctors develop more effective strategies for handling viral illnesses (Orlova et al.), (Aryan et al.).
| Technique | Resolution (nm) | Magnification | Sample Preparation | Cost | Applications |
| Light Microscopy | 200 | up to 2000x | Minimal | $1,000 – $50,000 | Living cells, Fungi, Bacteria |
| Electron Microscopy | 0.1 | up to 10,000,000x | Extensive (dehydration, staining) | $50,000 – $5 million | Viruses, Cellular structures, Protein complexes |
Microscopy Techniques Comparison
B. Bacterial Culture & Growth Media
Studying bacteria culture and growth media is key in microbiology. It helps scientists grow and manage microbial groups in controlled settings that can be carefully watched and changed. Different kinds of culture media, like selective, differential, and enriched media, are used to isolate certain bacteria or to encourage specific metabolic actions, which enables detailed examination of bacterial activities and biology under various situations. Selective media include certain substances that block unwanted bacteria but allow the target ones to grow. Differential media use indicators that change color based on certain metabolic actions, making it easier to identify bacteria based on their biochemical traits. Also, the growth media need to be tailored to match specific environmental factors, such as temperature and pH, so they can effectively imitate natural environments. This is crucial for understanding how bacteria behave in their natural settings. Recent studies stress the importance of using customized media to assess pathogen survival and responses to treatments, like analyzing how foodborne pathogens react to biocidal agents such as ozone, showing the important link between culture methods and assessments of microbial health (Anderson et al.). These methods not only improve our understanding of bacterial traits but also open doors for medical uses, particularly in fighting antibiotic resistance (Sahi et al.). Furthermore, ongoing developments in growth media designs, such as creating richer or more unique environments, continue to advance microbial research. An image illustrates this process through microbial diagnostics methods, underscoring the value of accurate culture techniques and the progress made in microbiological practices that enhance our capability to study and manipulate bacteria for various research and clinical needs.
| Media Type | Purpose | Common Use | Optimal PH | Storage Condition | Shelf Life |
| Nutrient Agar | General purpose growth of non-fibrous organisms | Isolated cultures in labs | 6.8 | Room temperature | 6 months |
| Mannitol Salt Agar | Selective for Staphylococci and differentiates Staphylococcus aureus | Clinical diagnostics | 7.4 | Room temperature | 6-12 months |
| MacConkey Agar | Isolates Gram-negative bacteria and differentiates lactose fermenters | Pathogen detection in samples | 7.1 | Room temperature | 4-6 months |
| Sabouraud Dextrose Agar | Supports growth of fungi and yeast | Fungal cultures in clinical settings | 5.6 | Refrigerated | 1 year |
| Blood Agar | Enrichment for fastidious organisms and differentiation of hemolytic activities | Bacterial identification and culture | 7.2 | Refrigerated | 1-2 weeks |
Bacterial Culture & Growth Media Statistics
C. DNA Sequencing and Genetic Analysis
DNA sequencing and genetic analysis change how we understand microbes and viruses. They offer important insights for research and clinical uses. Using new methods like next-generation sequencing (NGS) allows for thorough identification and study of pathogens, which helps us react to outbreaks and grasp disease processes better. Recent studies show that these genomic methods help find new viruses and play a big part in creating targeted treatments and vaccines, highlighting their role in public health efforts (Chiu et al.). Also, quickly sequencing genomes helps researchers watch for genetic changes in real-time, which is vital during epidemics when fast responses are needed. Furthermore, DNA sequencing technologies help uncover genetic differences among microorganism populations, identifying factors that cause disease and genes linked to antibiotic resistance (Anderson et al.). This skill is crucial now as antibiotic resistance rises, allowing for quick plans against resistant strains. In summary, these techniques are essential for advancing microbiological research and enhancing our ability to diagnose viruses, making DNA sequencing a key element in microbial study. As we keep delving into the details of microbial genomics, the impact of these technologies will surely grow, improving our understanding and control of infectious diseases and leading to new treatment options.
Here is a structured table on DNA Sequencing and Genetic Analysis:
| No. | Method/Concept | Description |
|---|---|---|
| 1 | Definition of DNA Sequencing | The process of determining the exact nucleotide order in a DNA molecule. |
| 2 | Sanger Sequencing | First-generation sequencing method using chain-termination with fluorescently labeled nucleotides. |
| 3 | Next-Generation Sequencing (NGS) | High-throughput sequencing allowing rapid genome analysis. |
| 4 | Whole Genome Sequencing (WGS) | Determines the complete DNA sequence of an organism’s genome. |
| 5 | Whole Exome Sequencing (WES) | Focuses on sequencing only the protein-coding regions (exons) of genes. |
| 6 | Third-Generation Sequencing | Includes technologies like PacBio SMRT and Oxford Nanopore, allowing real-time, long-read sequencing. |
| 7 | Single-Cell Sequencing | Analyzes the genome of individual cells, useful in cancer and microbial research. |
| 8 | Shotgun Sequencing | Randomly fragments DNA and sequences overlapping sections to reconstruct the genome. |
| 9 | RNA Sequencing (RNA-Seq) | Analyzes transcriptomes to study gene expression and regulation. |
| 10 | Metagenomic Sequencing | Studies genetic material directly from environmental samples to identify microbial communities. |
| 11 | Mitochondrial DNA Sequencing | Used for studying maternal lineage, evolutionary biology, and forensic science. |
| 12 | Epigenetic Analysis | Examines DNA methylation and histone modifications affecting gene expression. |
| 13 | Comparative Genomics | Compares DNA sequences across species to study evolutionary relationships. |
| 14 | CRISPR-Based Genome Editing | Uses CRISPR-Cas9 to modify DNA sequences for functional analysis and therapeutic applications. |
| 15 | Genome-Wide Association Studies (GWAS) | Identifies genetic variants associated with diseases by analyzing large populations. |
| 16 | Bioinformatics in Genetic Analysis | Uses computational tools to interpret sequencing data, identify mutations, and predict protein functions. |
| 17 | Phylogenetic Analysis | Uses DNA sequences to construct evolutionary trees and study species relationships. |
| 18 | Variant Calling | Identifies genetic mutations, such as SNPs and insertions/deletions, from sequencing data. |
| 19 | Functional Genomics | Studies how genes and genetic elements function within biological systems. |
| 20 | Personalized Medicine and Genomics | Uses genetic information to tailor medical treatments to individuals. |
III. Methods for Studying Viruses
In studying viruses, many methods have been created to help understand viral pathogens and their interactions with hosts. Techniques like polymerase chain reaction (PCR) and next-generation sequencing (NGS) have changed the way we detect and study viruses, enabling quick and accurate analysis of viral genomes. This is important for both diagnosing patients and researching outbreaks. These advances help scientists find not just known viruses but also new viral strains, adding to our understanding of virology. Moreover, immunological methods such as enzyme-linked immunosorbent assays (ELISA) are useful for measuring viral particles in different samples, which enhances our knowledge of viral load and how infections work. These tests are key in clinical environments for tracking viral infections and treatment effectiveness. An example of viral diagnostic methods shows the steps from sample collection to analysis, showcasing important progress in detection technologies in recent years. Additionally, to improve our understanding of how viruses and hosts interact, researchers also use cell culture systems. These systems offer insights into how viruses reproduce and cause disease, helping to explore how viruses enter cells and how the immune system reacts, which is essential for creating effective treatments. Such methods not only clarify viral behavior but also aid in developing vaccines and antiviral medications, playing a crucial role in public health (Bieniasz et al.), (Anderson et al.).
Here is a table outlining various Methods for Studying Viruses:
| No. | Method | Description |
|---|---|---|
| 1 | Plaque Assay | Measures viral infectivity by counting plaques (clear zones) in cell cultures. |
| 2 | Hemagglutination Assay (HA) | Detects viruses that agglutinate red blood cells, such as influenza virus. |
| 3 | Hemagglutination Inhibition Assay (HI) | Identifies virus-specific antibodies that prevent hemagglutination. |
| 4 | Enzyme-Linked Immunosorbent Assay (ELISA) | Detects viral antigens or antibodies using enzyme-linked antibodies. |
| 5 | Polymerase Chain Reaction (PCR) | Amplifies viral DNA/RNA for detection and quantification. |
| 6 | Reverse Transcription PCR (RT-PCR) | Converts viral RNA into cDNA before PCR amplification, used for RNA viruses. |
| 7 | Quantitative PCR (qPCR) | Measures viral load in real-time using fluorescent markers. |
| 8 | Next-Generation Sequencing (NGS) | Determines the complete viral genome for studying mutations and evolution. |
| 9 | Metagenomics | Identifies unknown viruses in clinical and environmental samples without prior knowledge of sequences. |
| 10 | Electron Microscopy (EM) | Provides high-resolution imaging of viral morphology. |
| 11 | X-ray Crystallography | Determines the atomic structure of viral proteins, aiding drug design. |
| 12 | Cryo-Electron Microscopy (Cryo-EM) | A high-resolution method for studying viral structures in near-native states. |
| 13 | Virus Culture in Cell Lines | Allows propagation and study of viruses in specific cell cultures. |
| 14 | Animal Models | Used to study viral pathogenesis and host immune responses. |
| 15 | Single-Cell RNA Sequencing (scRNA-seq) | Analyzes viral interactions at the single-cell level. |
| 16 | Flow Cytometry | Identifies virus-infected cells by labeling viral antigens or nucleic acids. |
| 17 | Western Blot | Detects viral proteins using antibody-based techniques. |
| 18 | CRISPR-Based Diagnostics | Uses CRISPR-Cas systems for rapid viral detection (e.g., SHERLOCK, DETECTR). |
| 19 | Serological Testing | Detects antibodies against viruses to assess immune response. |
| 20 | Bioinformatics and Computational Virology | Uses algorithms and databases to analyze viral evolution, epidemiology, and structural biology. |
A. Cell Cultures and Viral Growth Techniques
Cell cultures are very important in virology, helping to study how viruses act, how they replicate, and how they interact with host cells. These in vitro setups allow scientists to closely watch how viruses grow and are also crucial for developing vaccines and antiviral treatments. By using different culture methods, researchers can adjust important factors like nutrients, temperature, and pH to improve virus production and the results of their studies. Advanced molecular techniques improve these cultures, allowing for accurate identification and detailed analysis of viral strains, which helps to explain the complex ways viruses cause diseases. Furthermore, specific cell lines offer a controlled setting to investigate detailed viral-host interactions and can provide valuable information about how viruses cause illness, how the immune system responds, and potential treatment targets. Recent studies highlight how new techniques, such as using mycoviruses for biocontrol, show that ongoing improvements in cell culture methods are changing how we understand and tackle viral diseases globally (Anderson et al.), (Díez Casero et al.). The related image summarizes the significant changes in cell culture technologies, showing their major advancements, uses, and effects on current virology research. These advances are vital for understanding current viral risks and for anticipating and reducing future viral outbreaks, demonstrating the key role of cell cultures in virology and related fields.
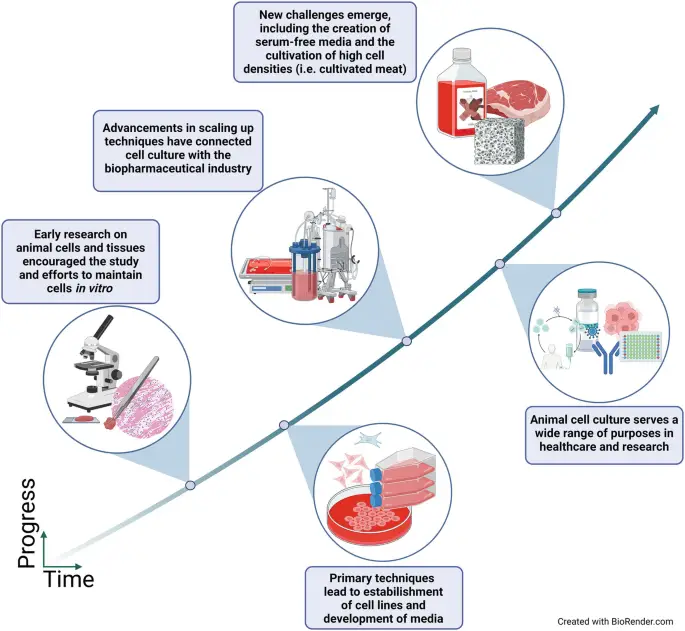
Image : Timeline of Advancements in Cell Culture Technology (The image illustrates the timeline of advancements in cell culture technology, highlighting key developments and challenges faced in the field. It begins with early research on animal cells and tissues, which laid the groundwork for maintaining cells in vitro. This is followed by significant progress in the establishment of cell lines and development of media. It also emphasizes advancements that connect cell culture with the biopharmaceutical industry, including the creation of serum-free media and high cell density cultivation. Overall, the infographic presents a concise overview of the evolution of cell culture practices and their implications for healthcare and research.)
B. PCR and Molecular Testing for Viruses
Polymerase Chain Reaction (PCR) and molecular testing have changed how we find and study viruses, being very important in both clinical and environmental areas. These methods allow for the identification of viral genomes with high sensitivity and accuracy, especially for viruses that cannot be grown in laboratories, like human caliciviruses (HuCVs) and small round-structured viruses (SRSVs) (Butcher et al.). The capability to find these hard-to-detect pathogens has really improved how public health responds and how surveillance systems work, ensuring outbreaks can be spotted and managed better. New improvements in PCR technology and the creation of in-depth molecular diagnostic methods have greatly expanded our knowledge of viral diversity and the complicated nature of viral diseases. These advancements not only shed light on viral mutations and evolution but also help monitor various viral pathogens that seriously affect agriculture, such as Grapevine leafroll-associated virus 3 (GLRaV-3) (Adib eRowhani et al.). By using molecular testing in farming, farmers and researchers can manage crop health better and reduce losses. A flowchart showing the diagnostic process in this paper simply outlines the PCR steps, explaining the method from sample collection to amplification and result interpretation. This flowchart is a crucial educational tool, assisting students, researchers, and practitioners in grasping the complexities of modern virology studies, ultimately promoting better public awareness and understanding of viral infections.
Here is a structured overview of PCR and Molecular Testing for Viruses in a tabular format:
| No. | Aspect | Description |
|---|---|---|
| 1 | Definition of Molecular Testing | Molecular testing involves detecting viral genetic material (DNA/RNA) using advanced techniques. |
| 2 | Polymerase Chain Reaction (PCR) | PCR is a widely used method to amplify and detect specific DNA or RNA sequences of viruses. |
| 3 | Types of PCR | Includes conventional PCR, real-time PCR (qPCR), and reverse transcription PCR (RT-PCR). |
| 4 | Reverse Transcription PCR (RT-PCR) | Converts viral RNA into complementary DNA (cDNA) before amplification, used for RNA viruses like SARS-CoV-2. |
| 5 | Quantitative PCR (qPCR) | Measures viral load in a sample using fluorescence-based detection methods. |
| 6 | Multiplex PCR | Detects multiple viral targets in a single reaction, useful in diagnosing co-infections. |
| 7 | Digital PCR (dPCR) | A highly sensitive technique providing absolute quantification of viral nucleic acids. |
| 8 | Loop-Mediated Isothermal Amplification (LAMP) | A rapid alternative to PCR that amplifies nucleic acids at a constant temperature. |
| 9 | Sample Collection | Nasopharyngeal swabs, throat swabs, saliva, and blood are commonly used for viral testing. |
| 10 | RNA Extraction | A crucial step where viral RNA is isolated before amplification in RT-PCR tests. |
| 11 | Target Gene Selection | PCR targets conserved viral genes like N, E, and RdRp in SARS-CoV-2. |
| 12 | Sensitivity and Specificity | PCR offers high sensitivity (detecting low viral loads) and high specificity (low false positives). |
| 13 | Turnaround Time | Results are typically available within a few hours to a day. |
| 14 | Use in Outbreaks | PCR is essential for early detection, contact tracing, and containment of viral outbreaks. |
| 15 | Comparison with Antigen Testing | PCR is more sensitive but slower than antigen-based rapid tests. |
| 16 | Automation in PCR Testing | Advanced platforms like RT-qPCR machines allow high-throughput viral testing. |
| 17 | Point-of-Care PCR | Portable PCR devices enable quick diagnosis in remote and resource-limited settings. |
| 18 | Challenges in PCR Testing | High costs, need for specialized equipment, and risk of contamination affecting results. |
| 19 | Future Trends | Integration of CRISPR-based diagnostics and AI-driven PCR analysis for faster and more accurate detection. |
| 20 | Global Impact | PCR remains the gold standard for viral diagnostics, essential in pandemic preparedness and surveillance. |
C. Cryo-EM and Viral Structure Studies
Cryo-electron microscopy (Cryo-EM) is an important tool for understanding viral structures closely, helping us learn more about viruses and their complexities. This method, which keeps biological samples very cold, lets scientists see viruses in their natural condition, providing valuable information about their structure and function. Recent research, like studies on the Ebola virus, shows how Cryo-EM can uncover detailed information about the arrangement of viral parts, such as glycoproteins and nucleocapsids. These components are vital for understanding how viruses cause diseases and escape immune responses, helping us see how viruses interact with their hosts (A Groseth et al.). Furthermore, research on hantaviruses using Cryo-EM has emphasized important structural features that aid in entering host cells, showing how antibodies can neutralize these viral threats. This gives us a useful model for understanding viral interactions and developing treatments, an area that is actively being researched (Bowden et al.). These developments highlight the importance of Cryo-EM in the search to use viral structures in making vaccines and antiviral medications, making it crucial for modern microbiology and virology. By providing clear and detailed images of viral particles, Cryo-EM has opened new possibilities in virology that were not possible before. The image that illustrates how Cryo-EM determines viral structures adds to this discussion, visually highlighting the complexities of the methods used and the significant potential of this imaging technique in uncovering the hidden details of viral life.
| Virus | Year | Resolution (Å) | Significance |
| SARS-CoV-2 | 2020 | 3.2 | Detailed understanding of spike protein structure for vaccine development. |
| Zika Virus | 2016 | 4 | Revealed structural changes that influence viral entry into host cells. |
| HIV | 2018 | 3.6 | Identified key structural features for potential therapeutic targets. |
| Ebola Virus | 2015 | 6.2 | Provided insights into viral assembly and infection mechanisms. |
| Influenza Virus | 2021 | 4.5 | Enhanced understanding of hemagglutinin and its role in virus entry. |
Cryo-EM Insights on Viral Structures
IV. The Role of Bioinformatics in Microbiology
Bioinformatics is very important in microbiology because it helps in analyzing and understanding large biological data from genomic sequencing, transcriptomics, and proteomics. The use of bioinformatics tools has changed how we look at microbial genomics, allowing scientists to find genetic differences and how different strains are related. These developments improve our understanding of microbial diversity and help identify specific genes associated with harmful traits, antibiotic resistance, and other key characteristics. With new high-throughput sequencing technologies, like those used in studying the African Swine Fever Virus (ASFV), researchers can explore the complicated relationships between hosts and pathogens, as shown in studies that illustrate how viral genes work with host immune systems (Costa et al.), (Blasdel et al.). Moreover, bioinformatics is vital in epidemiological research, where data mining and bioinformatics techniques help track the spread and evolution of infectious diseases, offering crucial information for public health measures. Also, bioinformatics supports the creation of personalized treatments and quick diagnostic tools, highlighting its role in fighting new infections. Using computational models to predict how microbes behave in different environments is another interesting aspect of bioinformatics, helping scientists to simulate possible outcomes of microbial interactions. A thorough understanding of these data-driven strategies is shown in image, which classifies various methods used for pathogen analysis, emphasizing the key role of bioinformatics in current microbiological research. Overall, as microbiology develops, the connection with bioinformatics will surely lead to new ideas and improve our capacity to tackle major issues in health and disease management.
| Application | Description | Impact | Source |
| Genomic Sequencing | Analyzing the genetic material of microbes to understand their capabilities and evolution. | Facilitates the identification of pathogenic organisms and antibiotic resistance genes. | National Center for Biotechnology Information (NCBI) |
| Protein Structure Prediction | Predicting the 3D structure of microbial proteins using computational methods. | Aids in drug design and understanding metabolic pathways. | Institute for Protein Design, University of Washington |
| Metagenomics | Studying genetic material recovered directly from environmental samples. | Provides insights into microbial diversity and ecosystem functions. | Microbiome Project |
| Phylogenetics | Analyzing evolutionary relationships among microbial species. | Helps in tracing the origins of infections and understanding microbial evolution. | Nature Reviews Microbiology |
| Database Management | Storing and managing large datasets of microbial genomes and proteomes. | Enables easy access to comprehensive microbial data for researchers. | European Nucleotide Archive (ENA) |
Bioinformatics Applications in Microbiology
A. How AI and Machine Learning Aid in Microbial Research
The use of artificial intelligence (AI) and machine learning (ML) in studying microbes is a big change in how we understand complicated relationships in microbial communities. This helps us learn more about science in this area. These tech methods allow for detailed reviews of large amounts of genomic and metabolomic data, which improves our ability to guess how microbes act and react to changes in their surroundings. For example, recent studies show that AI can find links between diet, the microbiome, and diseases like COVID-19, which shows more people are noticing how diet affects health ((Afkhami A et al.)). Additionally, events like the 2023 International Virus Bioinformatics Meeting show how these tools are changing today’s virology and microbiology studies, allowing for detailed looks at viral genomes and behaviors that we couldn’t analyze before ((Abecasis et al.)). By using AI and ML, scientists not only make microbial studies more accurate but also achieve major advances that can lead to better treatments. These new developments help us understand microbial activity better, which is key for dealing with important public health issues. The teamwork between data scientists and microbiologists supports a mixed approach that opens new paths for research and discovery in microbe studies, leading to new answers for difficult health problems. This partnership between technology and biology is essential for tackling the continuously changing problems posed by microbes and for creating plans that assure better health results for people everywhere.
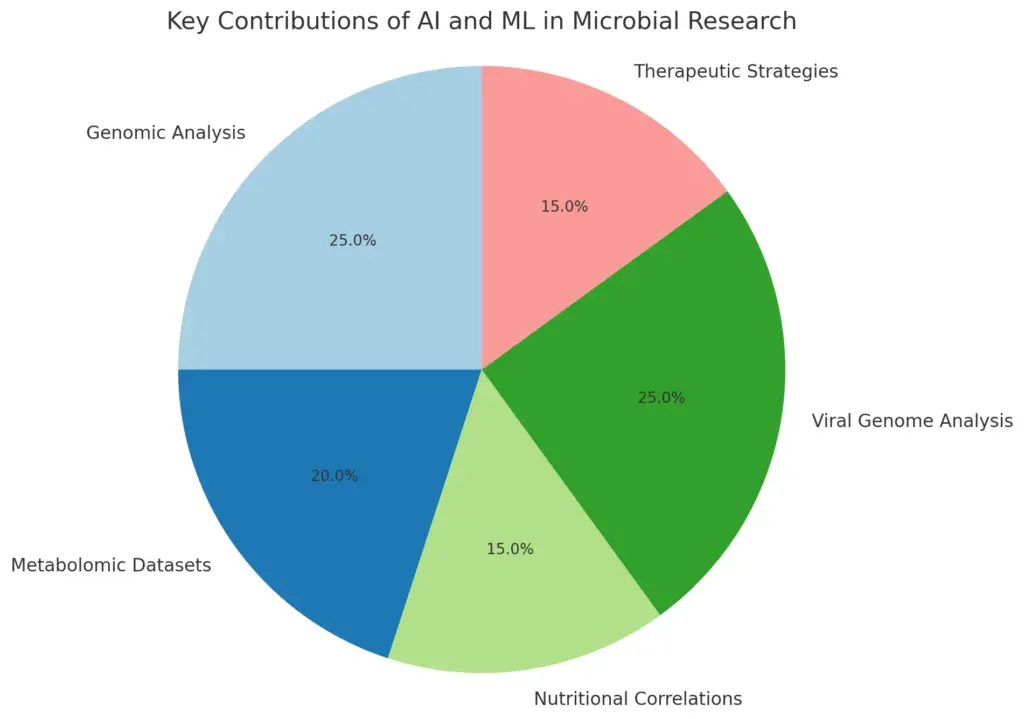
This pie chart represents the key contributions of AI and ML in microbial research, illustrating the relative focus on genomic analysis, metabolomic datasets, nutritional correlations, viral genome analysis, and the development of therapeutic strategies. Each section highlights the transformative role these technologies play in enhancing microbial understanding and public health outcomes.
B. Genome Sequencing and Virus Tracking
The growth of genome sequencing technology has changed how we track viral infections, helping us understand how viruses evolve and spread better. Methods like next-generation sequencing (NGS) make it possible to quickly and thoroughly sequence viral genomes. This helps detect genetic changes fast and gives important information about the behavior of viral quasispecies and mutations linked to drug resistance. For instance, many studies show how useful these modern methods are for profiling variability in human immunodeficiency virus (HIV) and hepatitis C virus (HCV), allowing doctors to create more targeted and effective treatment plans for these infections (Abbate et al.). Moreover, looking into the human virome with advanced metagenomic techniques uncovers the complex interactions of viruses in different ecosystems, which can greatly affect health, especially after lung transplants. This effect is clearly shown in various studies on lung transplant patients, where both the microbiome and virome are crucial to their overall health (Abbas et al.). These thorough methods highlight the essential role of genome sequencing in virology and related areas, leading to more effective public health responses and better management of viral outbreaks. Using these genomic technologies strengthens our ability to track new viral threats and improves our understanding of how viruses behave in real-time.
Here is a table with twenty key points on Genome Sequencing and Virus Tracking:
| No. | Key Point | Description |
|---|---|---|
| 1 | Definition of Genome Sequencing | Genome sequencing is the process of determining the complete DNA sequence of an organism’s genome. |
| 2 | Types of Genome Sequencing | Whole genome sequencing (WGS), whole exome sequencing (WES), and targeted sequencing are common types. |
| 3 | Next-Generation Sequencing (NGS) | NGS allows for rapid and high-throughput sequencing of viral genomes. |
| 4 | Role in Virus Identification | Genome sequencing helps in identifying and classifying new and existing viruses. |
| 5 | Mutation Detection | Sequencing enables the tracking of mutations in viral genomes, which can impact transmissibility and pathogenicity. |
| 6 | Epidemiological Tracking | Viral genome sequencing helps track the spread and evolution of viruses across populations and geographical locations. |
| 7 | Phylogenetic Analysis | Genome data is used to build phylogenetic trees to understand viral lineage relationships. |
| 8 | Role in Outbreak Investigations | Sequencing aids in identifying the source and transmission pathways of viral outbreaks. |
| 9 | SARS-CoV-2 Genome Sequencing | The COVID-19 pandemic showcased the importance of real-time genome sequencing in tracking variants. |
| 10 | Variants and Strains | Sequencing helps in detecting and categorizing new viral variants and subtypes. |
| 11 | Vaccine Development | Understanding viral genomes assists in designing effective vaccines. |
| 12 | Antiviral Drug Development | Genetic insights help in developing targeted antiviral therapies. |
| 13 | Public Health Surveillance | Sequencing enables monitoring of viruses for public health interventions. |
| 14 | Bioinformatics in Virus Tracking | Computational tools analyze and interpret large genomic datasets for virus tracking. |
| 15 | Metagenomic Sequencing | Identifies unknown viruses in clinical and environmental samples. |
| 16 | Global Sequencing Initiatives | Programs like GISAID and Nextstrain collect and share viral genome data worldwide. |
| 17 | Zoonotic Virus Detection | Genome sequencing helps in detecting viruses that jump from animals to humans (e.g., Ebola, Nipah). |
| 18 | Challenges in Sequencing | Cost, data analysis complexity, and sample quality can impact sequencing accuracy. |
| 19 | Ethical Considerations | Data privacy, biosecurity, and equitable access to sequencing technologies must be addressed. |
| 20 | Future of Genome Sequencing | Advances in AI and portable sequencing devices will enhance real-time virus tracking and response. |
V. Conclusion
To sum up, methods used in microbiology and virology have changed a lot, helping us understand and control pathogens better. This change is shown not just by old methods like culturing and microscopy but also by advanced molecular techniques that give us detailed information about viral genetics and how infections work. Recent research, like the study of PCV2 evolution and how it binds to receptors (Nauwynck et al.), shows the importance of new methods in monitoring genetic changes and figuring out how pathogens work. Also, the use of ozone for killing pathogens (Anderson et al.) shows new ways to improve food safety and public health. Helpful tools like the detailed diagnostic flowchart highlight the complicated and varied nature of current research techniques. All these improvements lead to a better understanding of how pathogens act, which is key for guiding public health measures and creating effective solutions.
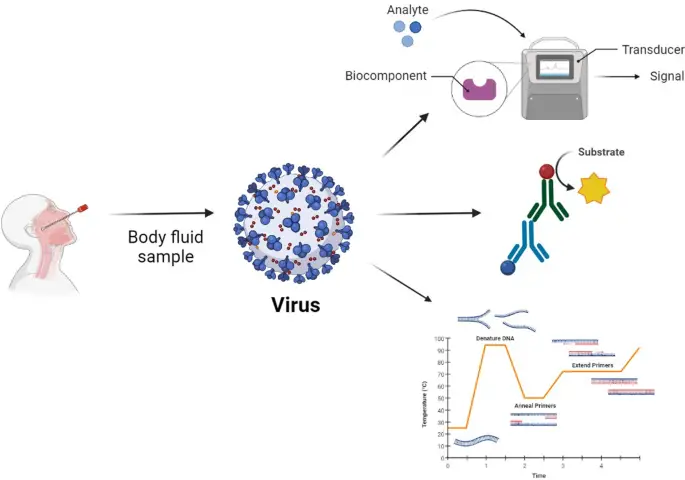
IMAGE – Diagnostic Process for Virus Detection from Body Fluid Samples (The image illustrates a diagnostic process for detecting viruses from body fluid samples. It depicts a human figure from which a sample is collected using a swab method. The virus is represented as a spherical structure with identifiable surface proteins. The diagram outlines the process flow, detailing the interaction between the analyte from the sample and a biocomponent within a transducer system, which converts the signal into a measurable output. Additionally, the image includes a lower section showing a temperature vs. time graph related to the polymerase chain reaction (PCR) process, indicating steps such as DNA denaturation, primer annealing, and extension. This visual synthesis of diagnostic methodology demonstrates protocols relevant to virology and molecular biology.)
A. How These Methods Help in Vaccine and Drug Development
The combining of different microbiological and virological methods is very important for faster vaccine and therapeutic drug development, changing modern medicine a lot. Methods like polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) are key, allowing for accurate detection and measurement of viruses, which helps us understand viral behavior and how to treat them better. These techniques reveal details about how viruses act, leading to better and more specific treatment options. Also, using peptide libraries shown on bacteriophage has changed how vaccines are designed, helping find mimotopes that are similar to natural antigens. This new method helps researchers create candidates that can trigger a more effective immune response. For instance, research showing the selection of dengue virus mimotopes highlights their ability to promote specific antibody creation, giving hope for improved vaccines against infectious diseases (Acosta et al.). Furthermore, methods like cell culture, along with molecular imaging, provide valuable information about how viruses act and how hosts interact, helping with drug-target identification and testing for safety and effectiveness. Overall, these advanced methods build a strong base that supports the development of new vaccines and drugs, allowing quicker reactions to health threats and improving our ability to fight infectious diseases worldwide. The combination of these methods not only speeds up research but also raises the chances of successful results in vaccine and drug development efforts.
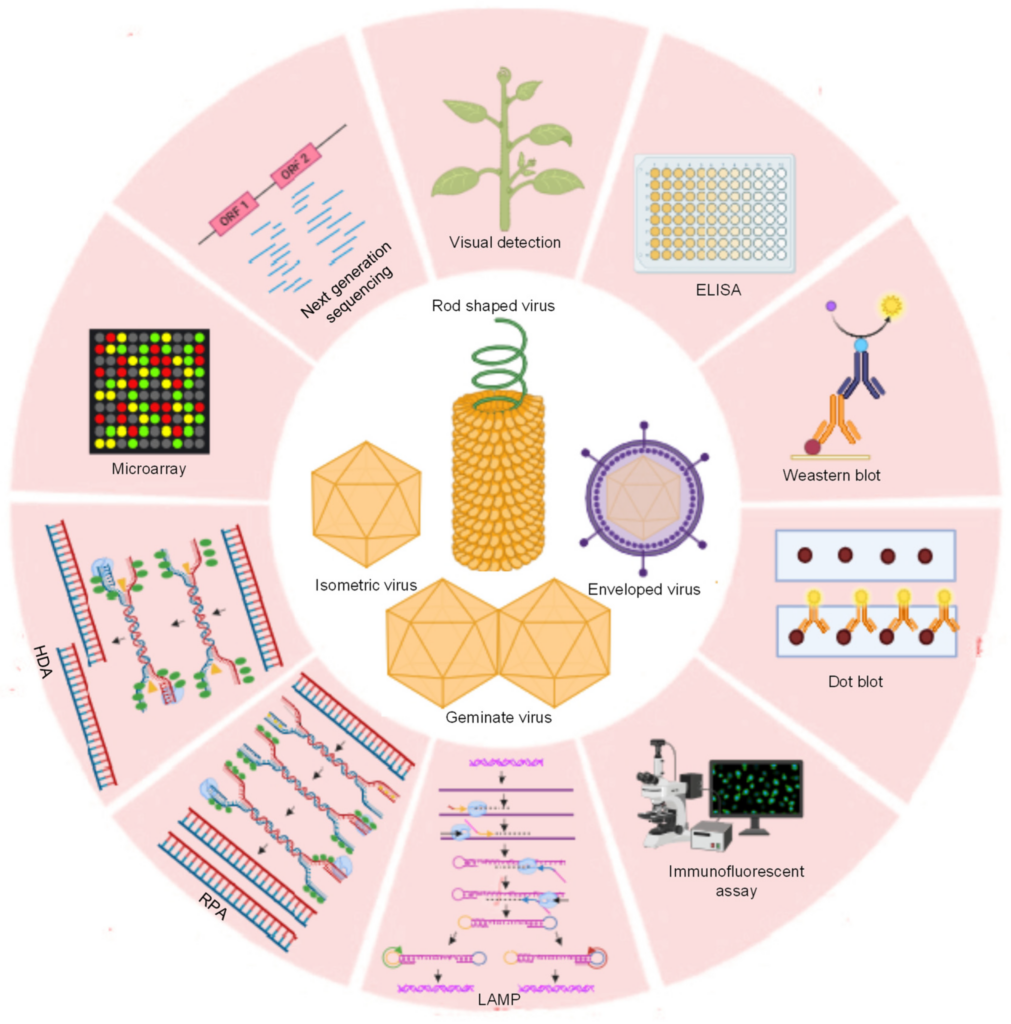
IMAGE – Diagram of Viral Structures and Molecular Detection Techniques (The image presents a circular diagram illustrating various molecular techniques and viral structures. Each segment features a distinct component related to virology and pathogen detection methods, including Next Generation Sequencing, ELISA, Western blot, and various types of viruses such as rod-shaped, isometric, and enveloped viruses. Additionally, the techniques highlighted include microarray analysis, LAMP, RPA, and immunofluorescent assays. This visual serves to integrate and summarize critical methodologies in viral research and diagnostics.)
B. The Future of Microbial Research
Microbial research is changing quickly and depends more on new methods and teamwork across different fields to better understand the complex interactions between microbes and their surroundings. Methods like next-generation sequencing and bioinformatics are key to finding the genetic basis of various microbial communities. This helps create stronger ecological models and identifies new species that are important for ecosystem health. Additionally, high-throughput screening and advanced imaging technologies allow researchers to see microbial activities in real time, offering new perspectives on their behaviors. Specifically, molecular techniques, as shown in [citeX], help clarify how viruses interact with host cells, providing valuable information about how diseases develop. These combined approaches are likely to lead future research in new ways, improving our capacity to tackle urgent global issues like emerging infectious diseases and antibiotic resistance. Furthermore, these advanced research methods will boost innovation in developing treatments and support efforts in environmental sustainability and bioremediation, aiming for healthier ecosystems and better public health. Looking ahead to the future of microbial research, there is great potential for significant discoveries, suggesting an exciting and promising field for scientific exploration.
REFERENCES
- Costa, Emanuel Nery de Oliveira Quartin, 1986-. “Viral modulation of interferon (IFN) responses to african swine fever virus (ASFV)”. 2011, https://core.ac.uk/download/12424877.pdf
- Blasdel, Bob G., Briers, Yves, Danis‐Wlodarczyk, Katarzyna, Drulis‐Kawa, et al.. “Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages”. ‘Wiley’, 2020, https://core.ac.uk/download/322827659.pdf
- Akbari Afkhami, Farhad, Ashkan, Mandana, Ghanbarzadeh, Erfan, Hedayati Ch, et al.. “New insight in severe acute respiratory syndrome coronavirus 2 consideration: Applied machine learning for nutrition quality, microbiome and microbial food poisoning concerns”. ParsPublisher, 2022, https://core.ac.uk/download/543100307.pdf
- Abecasis, Ana B, Babaian, Artem, Beck, Sebastian, Brierley, et al.. “The International Virus Bioinformatics Meeting 2023”. 2023, https://core.ac.uk/download/590313647.pdf
- Nauwynck, Hans, Theuns, Sebastiaan, Wei, Ruifang, Xie, et al.. “Changes on the viral capsid surface during the evolution of porcine circovirus type 2 (PCV2) from 2009 till 2018 may lead to a better receptor binding”. ‘Oxford University Press (OUP)’, 2019, https://core.ac.uk/download/225020523.pdf
- Anderson, J.G., Goudie, A.C., MacGregor, S.J., Olateju, et al.. “Inactivation of pathogens on food and contact surfaces using ozone as a biocidal agent”. 2003, https://core.ac.uk/download/9020578.pdf
- MetaSUB International Consortium. “The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium inaugural meeting report.”. eScholarship, University of California, 2016, https://core.ac.uk/download/323071097.pdf
- Butcher, S.A., Gould, E.A.. “Production of positive controls for calcivirus-specific PCR using recombinant baculiovirus technology”. ‘Freshwater Biological Association (BioOne sponsored)’, 1997, https://core.ac.uk/download/11020756.pdf
- Adib eRowhani, Anna E. C. Jooste, Daniel eCohen, Deborah A. Golino, Giovanni P. Martelli, Hans J. Maree, Hans J. Maree, et al.. “Grapevine leafroll-associated virus 3.”. eScholarship, University of California, 2013, https://core.ac.uk/download/323080750.pdf
- Bauer, J., Bornmann, L., Haunschild, R.. “Influential cited references in FEMS Microbiology Letters: lessons from Reference Publication Year Spectroscopy (RPYS)”. ‘Oxford University Press (OUP)’, 2019,
- Bieniasz, Paul D., Blanco-Melo, Daniel, Gifford, Robert J.. “Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors”. ‘eLife Sciences Publications, Ltd’, 2017, https://core.ac.uk/download/80696921.pdf
- Abbate, Allander, Allander, Ansorge, Archer, Barzon, Beerenwinkel, et al.. “Applications of Next-Generation Sequencing Technologies to Diagnostic Virology”. Molecular Diversity Preservation International (MDPI), 2011, https://core.ac.uk/download/pdf/8654683.pdf
- Abbas, Arwa. “The Human Lung Viral Microbiome In Health And Disease”. ScholarlyCommons, 2019, https://core.ac.uk/download/227276913.pdf
- Orlova, Elena. “Bacteriophages and their structural organisation”. ‘IntechOpen’, 2012, https://core.ac.uk/download/19509491.pdf
- Aryan, Ehsan, Fani, Mona, Gouklani, Hamed, Jalili, et al.. “What is the best laboratory method for diagnosis of Herpes Simplex Virus in genital infections?”. Publisher: Iranian Scientific Association of Clinical Laboratory, 2019, https://core.ac.uk/download/483639517.pdf
- A Groseth, A Sanchez, A Sanchez, B Heras, B Kastner, B Manicassamy, CL Hood, et al.. “The Organisation of Ebola Virus Reveals a Capacity for Extensive, Modular Polyploidy”. Public Library of Science, 2012, https://core.ac.uk/download/pdf/8668689.pdf
- Bowden, Thomas A., Doores, Katie J., Hepojoki, Jussi, Huiskonen, et al.. “Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein”. 2020, https://core.ac.uk/download/401690077.pdf
- Díez Casero, Julio Javier, Fernández Fernández, María Mercedes, Muñoz Adalia, Emigdio Jordán. “The use of mycoviruses in the control of forest diseases”. ‘Informa UK Limited’, 2016, https://core.ac.uk/download/323366858.pdf
- Acosta, Armando, Aguilar, Alicia, Alvarez, Mayling, Amin, et al.. “Immunogenicity of NS4b dengue 3 virus mimotope presented to the immune system as multiple antigen peptide system”. ‘Hindawi Limited’, 2013, https://core.ac.uk/download/19785886.pdf
- Sahi, Umar. “Isolation, characterization, and annotation of a novel mycobacteriophage Eaglepride”. 2022, https://core.ac.uk/download/534391870.pdf
- Chiu, Charles Y. “Viral pathogen discovery.”. eScholarship, University of California, 2013, https://core.ac.uk/download/323063718.pdf
Image References:
- “Timeline of the Evolution of Virology: Key Milestones and Technological Advances from the 1890s to the 2020s..” www.mdpi.com, 8 February 2025, https://www.mdpi.com/viruses/viruses-16-00374/article_deploy/html/images/viruses-16-00374-g001.png
- “Timeline of Advancements in Cell Culture Technology.” media.springernature.com, 8 February 2025, https://media.springernature.com/lw685/springer-static/image/chp%3A10.1007%2F978-3-031-55968-6_2/MediaObjects/604464_1_En_2_Figa_HTML.png
- “Diagnostic Process for Virus Detection from Body Fluid Samples.” media.springernature.com, 8 February 2025, https://media.springernature.com/lw685/springer-static/image/art%3A10.1186%2Fs12985-023-02108-w/MediaObjects/12985_2023_2108_Figa_HTML.png
- “Diagram of Viral Structures and Molecular Detection Techniques.” www.mdpi.com, 8 February 2025, https://www.mdpi.com/agriculture/agriculture-14-00284/article_deploy/html/images/agriculture-14-00284-g001.png
